Nascent chromatin capture proteomics determines chromatin dynamics during DNA replication and identifies unknown fork components
- PMID: 24561620
- PMCID: PMC4283098
- DOI: 10.1038/ncb2918
Nascent chromatin capture proteomics determines chromatin dynamics during DNA replication and identifies unknown fork components
Abstract
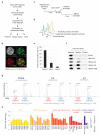
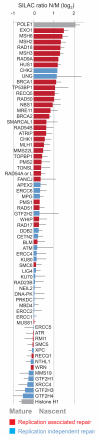
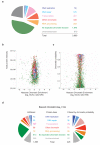
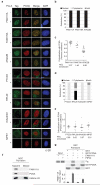
To maintain genome function and stability, DNA sequence and its organization into chromatin must be duplicated during cell division. Understanding how entire chromosomes are copied remains a major challenge. Here, we use nascent chromatin capture (NCC) to profile chromatin proteome dynamics during replication in human cells. NCC relies on biotin-dUTP labelling of replicating DNA, affinity purification and quantitative proteomics. Comparing nascent chromatin with mature post-replicative chromatin, we provide association dynamics for 3,995 proteins. The replication machinery and 485 chromatin factors such as CAF-1, DNMT1 and SUV39h1 are enriched in nascent chromatin, whereas 170 factors including histone H1, DNMT3, MBD1-3 and PRC1 show delayed association. This correlates with H4K5K12diAc removal and H3K9me1 accumulation, whereas H3K27me3 and H3K9me3 remain unchanged. Finally, we combine NCC enrichment with experimentally derived chromatin probabilities to predict a function in nascent chromatin for 93 uncharacterized proteins, and identify FAM111A as a replication factor required for PCNA loading. Together, this provides an extensive resource to understand genome and epigenome maintenance.
Figures



References
-
- Mechali M. Eukaryotic DNA replication origins: many choices for appropriate answers. Nat Rev Mol Cell Biol. 2010;11:728–738. - PubMed
-
- Alabert C, Groth A. Chromatin replication and epigenome maintenance. Nat Rev Mol Cell Biol. 2012;13:153–167. - PubMed
-
- Worcel A, Han S, Wong ML. Assembly of newly replicated chromatin. Cell. 1978;15:969–977. - PubMed
REFERENCES ONLINE METHODS
-
- Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. - PubMed
-
- Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc. 2006;1:2856–2860. - PubMed
-
- Rappsilber J, Ishihama Y, Mann M. Stop and go extraction tips for matrixassisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem. 2003;75:663–670. - PubMed
-
- Marzluff WF, Gongidi P, Woods KR, Jin J, Maltais LJ. The human and mouse replication-dependent histone genes. Genomics. 2002;80:487–498. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

