Tissue specific CTCF occupancy and boundary function at the human growth hormone locus
- PMID: 24561805
- PMCID: PMC4005687
- DOI: 10.1093/nar/gku139
Tissue specific CTCF occupancy and boundary function at the human growth hormone locus
Abstract
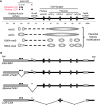
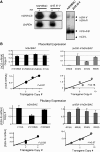
The robust and tissue-specific activation of the human growth hormone (hGH) gene cluster in the pituitary and placenta constitutes an informative model for analysis of gene regulation. The five-gene hGH cluster is regulated by two partially overlapping sets of DNase I hypersensitive sites (HSs) that constitute the pituitary (HSI, II, III and V) and placental (HSIII, IV, and V) locus control regions (LCRs). The single placenta-specific LCR component, HSIV, is located at -30 kb to the cluster. Here we generate a series of hGH/BAC transgenes specifically modified to identify structural features of the hGH locus required for its appropriate placental expression. We find that placental specificity is dependent on the overall multigene configuration of the cluster whereas the distance between the cluster and its LCR impacts the level of placental expression. We further observe that a major function of the placental hGH LCR is to insulate the transgene locus from site-of-integration effects. This insulation activity is linked to placenta-specific occupancy of the chromatin architectural protein, CTCF, at HSIV. These data reveal a remarkable combination of structural configurations and regulatory determinants that must work in concert to insure robust and tightly controlled expression from a complex multigene locus.
Figures






Similar articles
-
Evidence for Pituitary Repression of the Human Growth Hormone-Related Placental Lactogen Genes and a Role for P Sequences.Int J Mol Sci. 2025 May 6;26(9):4421. doi: 10.3390/ijms26094421. Int J Mol Sci. 2025. PMID: 40362658 Free PMC article. Review.
-
The human growth hormone gene cluster locus control region supports position-independent pituitary- and placenta-specific expression in the transgenic mouse.J Biol Chem. 2000 Mar 17;275(11):7902-9. doi: 10.1074/jbc.275.11.7902. J Biol Chem. 2000. PMID: 10713106
-
Patterns of histone acetylation suggest dual pathways for gene activation by a bifunctional locus control region.EMBO J. 2000 Dec 15;19(24):6814-22. doi: 10.1093/emboj/19.24.6814. EMBO J. 2000. PMID: 11118216 Free PMC article.
-
Enhancer-blocking activity is associated with hypersensitive site V sequences in the human growth hormone locus control region.DNA Cell Biol. 2011 Dec;30(12):995-1005. doi: 10.1089/dna.2011.1268. Epub 2011 Jun 28. DNA Cell Biol. 2011. PMID: 21711161
-
Obesity and regulation of human placental lactogen production in pregnancy.J Neuroendocrinol. 2020 Nov;32(11):e12859. doi: 10.1111/jne.12859. Epub 2020 Jun 5. J Neuroendocrinol. 2020. PMID: 32500948 Review.
Cited by
-
Long-range looping of a locus control region drives tissue-specific chromatin packing within a multigene cluster.Nucleic Acids Res. 2016 Jun 2;44(10):4651-64. doi: 10.1093/nar/gkw090. Epub 2016 Feb 17. Nucleic Acids Res. 2016. PMID: 26893355 Free PMC article.
-
Expression of Placental Members of the Human Growth Hormone Gene Family Is Increased in Response to Sequential Inhibition of DNA Methylation and Histone Deacetylation.Biores Open Access. 2015 Nov 1;4(1):446-56. doi: 10.1089/biores.2015.0036. eCollection 2015. Biores Open Access. 2015. PMID: 26634190 Free PMC article.
-
Tales from topographic oceans: topologically associated domains and cancer.Endocr Relat Cancer. 2019 Nov;26(11):R611-R626. doi: 10.1530/ERC-19-0348. Endocr Relat Cancer. 2019. PMID: 31505466 Free PMC article. Review.
-
Evidence for Pituitary Repression of the Human Growth Hormone-Related Placental Lactogen Genes and a Role for P Sequences.Int J Mol Sci. 2025 May 6;26(9):4421. doi: 10.3390/ijms26094421. Int J Mol Sci. 2025. PMID: 40362658 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

