Nanomechanics of β-rich proteins related to neuronal disorders studied by AFM, all-atom and coarse-grained MD methods
- PMID: 24562857
- PMCID: PMC3964301
- DOI: 10.1007/s00894-014-2144-5
Nanomechanics of β-rich proteins related to neuronal disorders studied by AFM, all-atom and coarse-grained MD methods
Abstract
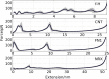
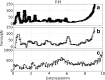
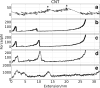
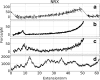
Computer simulations of protein unfolding substantially help to interpret force-extension curves measured in single-molecule atomic force microscope (AFM) experiments. Standard all-atom (AA) molecular dynamics simulations (MD) give a good qualitative mechanical unfolding picture but predict values too large for the maximum AFM forces with the common pulling speeds adopted here. Fine tuned coarse-grain MD computations (CG MD) offer quantitative agreement with experimental forces. In this paper we address an important methodological aspect of MD modeling, namely the impact of numerical noise generated by random assignments of bead velocities on maximum forces (F(max)) calculated within the CG MD approach. Distributions of CG forces from 2000 MD runs for several model proteins rich in β structures and having folds with increasing complexity are presented. It is shown that F(max) have nearly Gaussian distributions and that values of F(max) for each of those β-structures may vary from 93.2 ± 28.9 pN (neurexin) to 198.3 ± 25.2 pN (fibronectin). The CG unfolding spectra are compared with AA steered MD data and with results of our AFM experiments for modules present in contactin, fibronectin and neurexin. The stability of these proteins is critical for the proper functioning of neuronal synaptic clefts. Our results confirm that CG modeling of a single molecule unfolding is a good auxiliary tool in nanomechanics but large sets of data have to be collected before reliable comparisons of protein mechanical stabilities are made.
Figures






Similar articles
-
Nanomechanics of Ig-like domains of human contactin (BIG-2).J Mol Model. 2011 Sep;17(9):2313-23. doi: 10.1007/s00894-011-1010-y. Epub 2011 Mar 29. J Mol Model. 2011. PMID: 21445711 Free PMC article.
-
Reconstruction of mechanical unfolding and refolding pathways of proteins with atomic force spectroscopy and computer simulations.Methods. 2022 Jan;197:39-53. doi: 10.1016/j.ymeth.2021.05.012. Epub 2021 May 18. Methods. 2022. PMID: 34020035
-
Fully Atomistic Simulations of Protein Unfolding in Low Speed Atomic Force Microscope and Force Clamp Experiments with the Help of Boxed Molecular Dynamics.J Phys Chem B. 2016 Feb 4;120(4):700-8. doi: 10.1021/acs.jpcb.5b11519. Epub 2016 Jan 25. J Phys Chem B. 2016. PMID: 26760898
-
Experimental and computational characterization of biological liquid crystals: a review of single-molecule bioassays.Int J Mol Sci. 2009 Sep 10;10(9):4009-4032. doi: 10.3390/ijms10094009. Int J Mol Sci. 2009. PMID: 19865530 Free PMC article. Review.
-
Single molecule force spectroscopy using polyproteins.Chem Soc Rev. 2012 Jul 21;41(14):4781-96. doi: 10.1039/c2cs35033e. Epub 2012 May 30. Chem Soc Rev. 2012. PMID: 22648310 Review.
Cited by
-
Mathematical model of mechanobiology of acute and repeated synaptic injury and systemic biomarker kinetics.Front Cell Neurosci. 2023 Feb 6;17:1007062. doi: 10.3389/fncel.2023.1007062. eCollection 2023. Front Cell Neurosci. 2023. PMID: 36814869 Free PMC article.
-
Nanomechanics of multidomain neuronal cell adhesion protein contactin revealed by single molecule AFM and SMD.Sci Rep. 2017 Aug 18;7(1):8852. doi: 10.1038/s41598-017-09482-w. Sci Rep. 2017. PMID: 28821864 Free PMC article.
-
Dynamics, nanomechanics and signal transduction in reelin repeats.Sci Rep. 2019 Dec 12;9(1):18974. doi: 10.1038/s41598-019-55461-8. Sci Rep. 2019. PMID: 31831824 Free PMC article.
-
Entropic bonding of the type 1 pilus from experiment and simulation.R Soc Open Sci. 2020 Apr 15;7(4):200183. doi: 10.1098/rsos.200183. eCollection 2020 Apr. R Soc Open Sci. 2020. PMID: 32431906 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

