Genetic validation of aminoacyl-tRNA synthetases as drug targets in Trypanosoma brucei
- PMID: 24562907
- PMCID: PMC4000095
- DOI: 10.1128/EC.00017-14
Genetic validation of aminoacyl-tRNA synthetases as drug targets in Trypanosoma brucei
Abstract
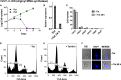
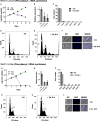
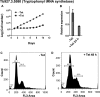
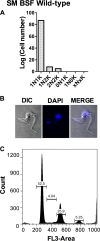
Human African trypanosomiasis (HAT) is an important public health threat in sub-Saharan Africa. Current drugs are unsatisfactory, and new drugs are being sought. Few validated enzyme targets are available to support drug discovery efforts, so our goal was to obtain essentiality data on genes with proven utility as drug targets. Aminoacyl-tRNA synthetases (aaRSs) are known drug targets for bacterial and fungal pathogens and are required for protein synthesis. Here we survey the essentiality of eight Trypanosoma brucei aaRSs by RNA interference (RNAi) gene expression knockdown, covering an enzyme from each major aaRS class: valyl-tRNA synthetase (ValRS) (class Ia), tryptophanyl-tRNA synthetase (TrpRS-1) (class Ib), arginyl-tRNA synthetase (ArgRS) (class Ic), glutamyl-tRNA synthetase (GluRS) (class 1c), threonyl-tRNA synthetase (ThrRS) (class IIa), asparaginyl-tRNA synthetase (AsnRS) (class IIb), and phenylalanyl-tRNA synthetase (α and β) (PheRS) (class IIc). Knockdown of mRNA encoding these enzymes in T. brucei mammalian stage parasites showed that all were essential for parasite growth and survival in vitro. The reduced expression resulted in growth, morphological, cell cycle, and DNA content abnormalities. ThrRS was characterized in greater detail, showing that the purified recombinant enzyme displayed ThrRS activity and that the protein localized to both the cytosol and mitochondrion. Borrelidin, a known inhibitor of ThrRS, was an inhibitor of T. brucei ThrRS and showed antitrypanosomal activity. The data show that aaRSs are essential for T. brucei survival and are likely to be excellent targets for drug discovery efforts.
Figures






Similar articles
-
Genetic manipulation of Leishmania donovani threonyl tRNA synthetase facilitates its exploration as a potential therapeutic target.PLoS Negl Trop Dis. 2018 Jun 13;12(6):e0006575. doi: 10.1371/journal.pntd.0006575. eCollection 2018 Jun. PLoS Negl Trop Dis. 2018. PMID: 29897900 Free PMC article.
-
Non-canonical eukaryotic glutaminyl- and glutamyl-tRNA synthetases form mitochondrial aminoacyl-tRNA in Trypanosoma brucei.J Biol Chem. 2004 Jan 9;279(2):1161-6. doi: 10.1074/jbc.M310100200. Epub 2003 Oct 16. J Biol Chem. 2004. PMID: 14563839
-
Aminoacyl tRNA synthetases as malarial drug targets: a comparative bioinformatics study.Malar J. 2019 Feb 6;18(1):34. doi: 10.1186/s12936-019-2665-6. Malar J. 2019. PMID: 30728021 Free PMC article.
-
Aminoacyl-tRNA Synthetases: On Anti-Synthetase Syndrome and Beyond.Front Immunol. 2022 May 13;13:866087. doi: 10.3389/fimmu.2022.866087. eCollection 2022. Front Immunol. 2022. PMID: 35634293 Free PMC article. Review.
-
Structural analyses of the malaria parasite aminoacyl-tRNA synthetases provide new avenues for antimalarial drug discovery.Protein Sci. 2021 Sep;30(9):1793-1803. doi: 10.1002/pro.4148. Protein Sci. 2021. PMID: 34184352 Free PMC article. Review.
Cited by
-
A binding hotspot in Trypanosoma cruzi histidyl-tRNA synthetase revealed by fragment-based crystallographic cocktail screens.Acta Crystallogr D Biol Crystallogr. 2015 Aug;71(Pt 8):1684-98. doi: 10.1107/S1399004715007683. Epub 2015 Jul 31. Acta Crystallogr D Biol Crystallogr. 2015. PMID: 26249349 Free PMC article.
-
Genome-scale RNAi screens for high-throughput phenotyping in bloodstream-form African trypanosomes.Nat Protoc. 2015 Jan;10(1):106-33. doi: 10.1038/nprot.2015.005. Epub 2014 Dec 11. Nat Protoc. 2015. PMID: 25502887
-
Genetic Validation of Leishmania donovani Lysyl-tRNA Synthetase Shows that It Is Indispensable for Parasite Growth and Infectivity.mSphere. 2017 Aug 30;2(4):e00340-17. doi: 10.1128/mSphereDirect.00340-17. eCollection 2017 Jul-Aug. mSphere. 2017. PMID: 28875178 Free PMC article.
-
Anti-trypanosomatid drug discovery: an ongoing challenge and a continuing need.Nat Rev Microbiol. 2017 Feb 27;15(4):217-231. doi: 10.1038/nrmicro.2016.193. Online ahead of print. Nat Rev Microbiol. 2017. PMID: 28239154 Free PMC article. Review.
-
Novel Effects of Lapatinib Revealed in the African Trypanosome by Using Hypothesis-Generating Proteomics and Chemical Biology Strategies.Antimicrob Agents Chemother. 2017 Jan 24;61(2):e01865-16. doi: 10.1128/AAC.01865-16. Print 2017 Feb. Antimicrob Agents Chemother. 2017. PMID: 27872081 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

