The Entamoeba histolytica Dnmt2 homolog (Ehmeth) confers resistance to nitrosative stress
- PMID: 24562908
- PMCID: PMC4000097
- DOI: 10.1128/EC.00031-14
The Entamoeba histolytica Dnmt2 homolog (Ehmeth) confers resistance to nitrosative stress
Abstract
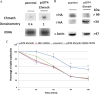
Nitric oxide (NO) has antimicrobial properties against many pathogens due to its reactivity as an S-nitrosylating agent. It inhibits many of the key enzymes that are involved in the metabolism and virulence of the parasite Entamoeba histolytica through S-nitrosylation of essential cysteine residues. Very little information is available on the mechanism of resistance to NO by pathogens in general and by this parasite in particular. Here, we report that exposure of the parasites to S-nitrosoglutathione (GSNO), an NO donor molecule, strongly reduces their viability and protein synthesis. However, the deleterious effects of NO were significantly reduced in trophozoites overexpressing Ehmeth, the cytosine-5 methyltransferase of the Dnmt2 family. Since these trophozoites also exhibited high levels of tRNA(Asp) methylation, the high levels suggested that Ehmeth-mediated tRNA(Asp) methylation is part of the resistance mechanism to NO. We previously reported that enolase, another glycolytic enzyme, binds to Ehmeth and inhibits its activity. We observed that the amount of Ehmeth-enolase complex was significantly reduced in GSNO-treated E. histolytica, which explains the aforementioned increase of tRNA methylation. Specifically, we demonstrated via site-directed mutagenesis that cysteine residues 228 and 229 of Ehmeth are susceptible to S-nitrosylation and are crucial for Ehmeth binding to enolase and for Ehmeth-mediated resistance to NO. These results indicate that Ehmeth has a central role in the response of the parasite to NO, and they contribute to the growing evidence that NO is a regulator of epigenetic mechanisms.
Figures


Similar articles
-
A new nuclear function of the Entamoeba histolytica glycolytic enzyme enolase: the metabolic regulation of cytosine-5 methyltransferase 2 (Dnmt2) activity.PLoS Pathog. 2010 Feb 19;6(2):e1000775. doi: 10.1371/journal.ppat.1000775. PLoS Pathog. 2010. PMID: 20174608 Free PMC article.
-
In vitro tRNA methylation assay with the Entamoeba histolytica DNA and tRNA methyltransferase Dnmt2 (Ehmeth) enzyme.J Vis Exp. 2010 Oct 19;(44):2390. doi: 10.3791/2390. J Vis Exp. 2010. PMID: 21048666 Free PMC article.
-
Characterization of cytosine methylated regions and 5-cytosine DNA methyltransferase (Ehmeth) in the protozoan parasite Entamoeba histolytica.Nucleic Acids Res. 2004 Jan 9;32(1):287-97. doi: 10.1093/nar/gkh161. Print 2004. Nucleic Acids Res. 2004. PMID: 14715927 Free PMC article.
-
Lipids in Entamoeba histolytica: Host-Dependence and Virulence Factors.Front Cell Infect Microbiol. 2020 Mar 10;10:75. doi: 10.3389/fcimb.2020.00075. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 32211340 Free PMC article. Review.
-
Trophozoites of Entamoeba histolytica epigenetically silenced in several genes are virulence-attenuated.Parasite. 2008 Sep;15(3):266-74. doi: 10.1051/parasite/2008153266. Parasite. 2008. PMID: 18814693 Review.
Cited by
-
Exploring the Interactome of the Queuine Salvage Protein DUF2419 in Entamoeba histolytica.Cells. 2024 Nov 18;13(22):1900. doi: 10.3390/cells13221900. Cells. 2024. PMID: 39594649 Free PMC article.
-
RNA methylation and cellular response to oxidative stress-promoting anticancer agents.Cell Cycle. 2023 Apr;22(8):870-905. doi: 10.1080/15384101.2023.2165632. Epub 2023 Jan 17. Cell Cycle. 2023. PMID: 36648057 Free PMC article. Review.
-
Reviving the RNA World: An Insight into the Appearance of RNA Methyltransferases.Front Genet. 2016 Jun 6;7:99. doi: 10.3389/fgene.2016.00099. eCollection 2016. Front Genet. 2016. PMID: 27375676 Free PMC article. Review.
-
Characterization of Cytosine Methylation and the DNA Methyltransferases of Toxoplasma gondii.Int J Biol Sci. 2017 Mar 11;13(4):458-470. doi: 10.7150/ijbs.18644. eCollection 2017. Int J Biol Sci. 2017. PMID: 28529454 Free PMC article.
-
Proteomic Identification of Oxidized Proteins in Entamoeba histolytica by Resin-Assisted Capture: Insights into the Role of Arginase in Resistance to Oxidative Stress.PLoS Negl Trop Dis. 2016 Jan 6;10(1):e0004340. doi: 10.1371/journal.pntd.0004340. eCollection 2016 Jan. PLoS Negl Trop Dis. 2016. PMID: 26735309 Free PMC article.
References
-
- . 1997. WHO/PAHO/UNESCO report. A consultation with experts on amoebiasis. Mexico City, Mexico 28–29 January, 1997. Epidemiol. Bull. 18:13–14 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

