Escherichia coli isolate for studying colonization of the mouse intestine and its application to two-component signaling knockouts
- PMID: 24563035
- PMCID: PMC3993324
- DOI: 10.1128/JB.01296-13
Escherichia coli isolate for studying colonization of the mouse intestine and its application to two-component signaling knockouts
Abstract
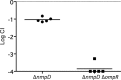
The biology of Escherichia coli in its primary niche, the animal intestinal tract, is remarkably unexplored. Studies with the streptomycin-treated mouse model have produced important insights into the metabolic requirements for Escherichia coli to colonize mice. However, we still know relatively little about the physiology of this bacterium growing in the complex environment of an intestine that is permissive for the growth of competing flora. We have developed a system for studying colonization using an E. coli strain, MP1, isolated from a mouse. MP1 is genetically tractable and does not require continuous antibiotic treatment for stable colonization. As an application of this system, we separately knocked out each two-component system response regulator in MP1 and performed competitions against the wild-type strain. We found that only three response regulators, ArcA, CpxR, and RcsB, produce strong colonization defects, suggesting that in addition to anaerobiosis, adaptation to cell envelope stress is a critical requirement for E. coli colonization of the mouse intestine. We also show that the response regulator OmpR, which had previously been hypothesized to be important for adaptation between in vivo and ex vivo environments, is not required for MP1 colonization due to the presence of a third major porin.
Figures






Similar articles
-
An Escherichia coli Nissle 1917 missense mutant colonizes the streptomycin-treated mouse intestine better than the wild type but is not a better probiotic.Infect Immun. 2014 Feb;82(2):670-82. doi: 10.1128/IAI.01149-13. Epub 2013 Nov 25. Infect Immun. 2014. PMID: 24478082 Free PMC article.
-
Microbial Cell Factories à la Carte: Elimination of Global Regulators Cra and ArcA Generates Metabolic Backgrounds Suitable for the Synthesis of Bioproducts in Escherichia coli.Appl Environ Microbiol. 2018 Sep 17;84(19):e01337-18. doi: 10.1128/AEM.01337-18. Print 2018 Oct 1. Appl Environ Microbiol. 2018. PMID: 30030227 Free PMC article.
-
The Escherichia coli CpxA-CpxR envelope stress response system regulates expression of the porins ompF and ompC.J Bacteriol. 2005 Aug;187(16):5723-31. doi: 10.1128/JB.187.16.5723-5731.2005. J Bacteriol. 2005. PMID: 16077119 Free PMC article.
-
Cellular and molecular physiology of Escherichia coli in the adaptation to aerobic environments.J Biochem. 1996 Dec;120(6):1055-63. doi: 10.1093/oxfordjournals.jbchem.a021519. J Biochem. 1996. PMID: 9010748 Review.
-
Adaptation of Escherichia coli to redox environments by gene expression.Mol Microbiol. 1993 Jul;9(1):9-15. doi: 10.1111/j.1365-2958.1993.tb01664.x. Mol Microbiol. 1993. PMID: 8412675 Review.
Cited by
-
Oxygen-Dependent Cell-to-Cell Variability in the Output of the Escherichia coli Tor Phosphorelay.J Bacteriol. 2015 Jun 15;197(12):1976-87. doi: 10.1128/JB.00074-15. Epub 2015 Mar 30. J Bacteriol. 2015. PMID: 25825431 Free PMC article.
-
Siderophore piracy enhances Vibrio cholerae environmental survival and pathogenesis.Microbiology (Reading). 2020 Nov;166(11):1038-1046. doi: 10.1099/mic.0.000975. Epub 2020 Oct 5. Microbiology (Reading). 2020. PMID: 33074088 Free PMC article.
-
Isolation and Characterization of a Novel Temperate Escherichia coli Bacteriophage, Kapi1, Which Modifies the O-Antigen and Contributes to the Competitiveness of Its Host during Colonization of the Murine Gastrointestinal Tract.mBio. 2022 Feb 22;13(1):e0208521. doi: 10.1128/mbio.02085-21. Epub 2022 Jan 25. mBio. 2022. PMID: 35073745 Free PMC article.
-
Deciphering the Role of Colicins during Colonization of the Mammalian Gut by Commensal E. coli.Microorganisms. 2020 May 2;8(5):664. doi: 10.3390/microorganisms8050664. Microorganisms. 2020. PMID: 32370119 Free PMC article.
-
The Influence of Topinambur and Inulin Preventive Supplementation on Microbiota, Anxious Behavior, Cognitive Functions and Neurogenesis in Mice Exposed to the Chronic Unpredictable Mild Stress.Nutrients. 2023 Apr 23;15(9):2041. doi: 10.3390/nu15092041. Nutrients. 2023. PMID: 37432210 Free PMC article.
References
-
- Gordon DM, FitzGibbon F. 1999. The distribution of enteric bacteria from Australian mammals: host and geographical effects. Microbiology 145:2663–2671 - PubMed
-
- Rasko DA, Rosovitz MJ, Myers GS, Mongodin EF, Fricke WF, Gajer P, Crabtree J, Sebaihia M, Thomson NR, Chaudhuri R, Henderson IR, Sperandio V, Ravel J. 2008. The pangenome structure of Escherichia coli: comparative genomic analysis of E. coli commensal and pathogenic isolates. J. Bacteriol. 190:6881–6893. 10.1128/JB.00619-08 - DOI - PMC - PubMed
-
- Touchon M, Hoede C, Tenaillon O, Barbe V, Baeriswyl S, Bidet P, Bingen E, Bonacorsi S, Bouchier C, Bouvet O, Calteau A, Chiapello H, Clermont O, Cruveiller S, Danchin A, Diard M, Dossat C, Karoui ME, Frapy E, Garry L, Ghigo JM, Gilles AM, Johnson J, Le Bouguenec C, Lescat M, Mangenot S, Martinez-Jehanne V, Matic I, Nassif X, Oztas S, Petit MA, Pichon C, Rouy Z, Ruf CS, Schneider D, Tourret J, Vacherie B, Vallenet D, Medigue C, Rocha EP, Denamur E. 2009. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet. 5:e1000344. 10.1371/journal.pgen.1000344 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

