Profiling Gene Expression in Germinating Brassica Roots
- PMID: 24563578
- PMCID: PMC3926982
- DOI: 10.1007/s11105-013-0668-y
Profiling Gene Expression in Germinating Brassica Roots
Abstract
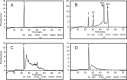
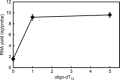
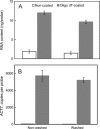
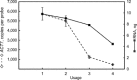
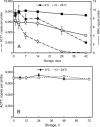
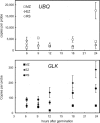
Based on previously developed solid-phase gene extraction (SPGE) we examined the mRNA profile in primary roots of Brassica rapa seedlings for highly expressed genes like ACT7 (actin7), TUB (tubulin1), UBQ (ubiquitin), and low expressed GLK (glucokinase) during the first day post-germination. The assessment was based on the mRNA load of the SPGE probe of about 2.1 ng. The number of copies of the investigated genes changed spatially along the length of primary roots. The expression level of all genes differed significantly at each sample position. Among the examined genes ACT7 expression was most even along the root. UBQ was highest at the tip and root-shoot junction (RS). TUB and GLK showed a basipetal gradient. The temporal expression of UBQ was highest in the MZ 9 h after primary root emergence and higher than at any other sample position. Expressions of GLK in EZ and RS increased gradually over time. SPGE extraction is the result of oligo-dT and oligo-dA hybridization and the results illustrate that SPGE can be used for gene expression profiling at high spatial and temporal resolution. SPGE needles can be used within two weeks when stored at 4 °C. Our data indicate that gene expression studies that are based on the entire root miss important differences in gene expression that SPGE is able to resolve for example growth adjustments during gravitropism.
Keywords: Brassica; Gene expression profiling; Solid-phase gene extraction; mRNA quality.
Figures

Similar articles
-
Transcription Profile of Auxin Related Genes during Positively Gravitropic Hypocotyl Curvature of Brassica rapa.Plants (Basel). 2022 Apr 28;11(9):1191. doi: 10.3390/plants11091191. Plants (Basel). 2022. PMID: 35567192 Free PMC article.
-
Augmentation of root gravitropism by hypocotyl curvature in Brassica rapa seedlings.Plant Sci. 2019 Aug;285:214-223. doi: 10.1016/j.plantsci.2019.05.017. Epub 2019 May 25. Plant Sci. 2019. PMID: 31203886
-
Solid phase gene extraction isolates mRNA at high spatial and temporal resolution.Biotechniques. 2008 Aug;45(2):172-8. doi: 10.2144/000112831. Biotechniques. 2008. PMID: 18687066
-
Assessing Radish Health during Space Cultivation by Gene Transcription.Plants (Basel). 2023 Sep 30;12(19):3458. doi: 10.3390/plants12193458. Plants (Basel). 2023. PMID: 37836197 Free PMC article.
-
Differential growth and hormone redistribution in gravireacting maize roots.Environ Exp Bot. 1989 Jan;29(1):37-45. doi: 10.1016/0098-8472(89)90037-3. Environ Exp Bot. 1989. PMID: 11541034 Review.
Cited by
-
Integration of Transcriptomic and Proteomic Profiles Reveals Multiple Levels of Genetic Regulation of Taproot Growth in Sugar Beet (Beta vulgaris L.).Front Plant Sci. 2022 Jul 13;13:882753. doi: 10.3389/fpls.2022.882753. eCollection 2022. Front Plant Sci. 2022. PMID: 35909753 Free PMC article.
-
Transcription Profile of Auxin Related Genes during Positively Gravitropic Hypocotyl Curvature of Brassica rapa.Plants (Basel). 2022 Apr 28;11(9):1191. doi: 10.3390/plants11091191. Plants (Basel). 2022. PMID: 35567192 Free PMC article.
-
Temperature Regulation of Primary and Secondary Seed Dormancy in Rosa canina L.: Findings from Proteomic Analysis.Int J Mol Sci. 2020 Sep 23;21(19):7008. doi: 10.3390/ijms21197008. Int J Mol Sci. 2020. PMID: 32977616 Free PMC article.
-
Transcriptome Profiling of Taproot Reveals Complex Regulatory Networks during Taproot Thickening in Radish (Raphanus sativus L.).Front Plant Sci. 2016 Aug 22;7:1210. doi: 10.3389/fpls.2016.01210. eCollection 2016. Front Plant Sci. 2016. PMID: 27597853 Free PMC article.
-
RNA capture pin technology: investigating long-term stability and mRNA purification specificity of oligonucleotide immobilization on gold and streptavidin surfaces.Anal Bioanal Chem. 2023 Oct;415(24):6077-6089. doi: 10.1007/s00216-023-04882-6. Epub 2023 Jul 29. Anal Bioanal Chem. 2023. PMID: 37516691
References
-
- Baluška F, Vitha S, Barlow PW, Volkmann D. Rearrangements of F‑actin arrays in growing cells of intact maize root apex tissues: a major developmental switch occurs in the postmitotic transition region. Eur J Cell Biol. 1997;72:113–121. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
