Chromatin redistribution of the DEK oncoprotein represses hTERT transcription in leukemias
- PMID: 24563617
- PMCID: PMC3927101
- DOI: 10.1593/neo.131658
Chromatin redistribution of the DEK oncoprotein represses hTERT transcription in leukemias
Abstract
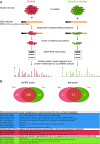
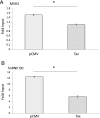
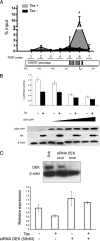
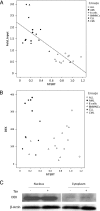
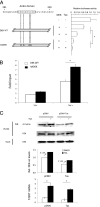
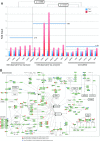
Although numerous factors have been found to modulate hTERT transcription, the mechanism of its repression in certain leukemias remains unknown. We show here that DEK represses hTERT transcription through its enrichment on the hTERT promoter in cells from chronic and acute myeloid leukemias, chronic lymphocytic leukemia, but not acute lymphocytic leukemias where hTERT is overexpressed. We isolated DEK from the hTERT promoter incubated with nuclear extracts derived from fresh acute myelogenous leukemia (AML) cells and from cells expressing Tax, an hTERT repressor encoded by the human T cell leukemia virus type 1. In addition to the recruitment of DEK, the displacement of two potent known hTERT transactivators from the hTERT promoter characterized both AML cells and Tax-expressing cells. Reporter and chromatin immunoprecipitation assays permitted to map the region that supports the repressive effect of DEK on hTERT transcription, which was proportionate to the level of DEK-promoter association but not with the level of DEK expression. Besides hTERT repression, this context of chromatin redistribution of DEK was found to govern about 40% of overall transcriptional modifications, including those of cancer-prone genes. In conclusion, DEK emerges as an hTERT repressor shared by various leukemia subtypes and seems involved in the deregulation of numerous genes associated with leukemogenesis.
Figures
References
-
- Campbell LJ, Fidler C, Eagleton H, Peniket A, Kusec R, Gal S, Littlewood TJ, Wainscoat JS, Boultwood J. hTERT, the catalytic component of telomerase, is downregulated in the haematopoietic stem cells of patients with chronic myeloid leukaemia. Leukemia. 2006;20:671–679. - PubMed
-
- Drummond MW, Hoare SF, Monaghan A, Graham SM, Alcorn MJ, Keith WN, Holyoake TL. Dysregulated expression of the major telomerase components in leukaemic stem cells. Leukemia. 2005;19:381–389. - PubMed
Supplementary References
-
- Gabet AS, Mortreux F, Charneau P, Riou P, Duc-Dodon M, Wu Y, Jeang KT, Wattel E. Inactivation of hTERT transcription by Tax. Oncogene. 2003;22(24):3734–3741. Prepublished on 2003/06/13 as doi:10.1038/sj.onc.1206468. - PubMed
-
- Hausmann S, Biddison WE, Smith KJ, Ding YH, Garboczi DN, Utz U, Wiley DC, Wucherpfennig KW. Peptide recognition by two HLA-A2/Tax11-19-specific T cell clones in relationship to theirMHC/peptide/TCR crystal structures. J Immunol. 1999;162(9):5389–5397. - PubMed
-
- Cleary J, Sitwala KV, Khodadoust MS, Kwok RP, Mor-Vaknin N, Cebrat M, Cole PA, Markovitz DM. p300/CBP-associated factor drives DEK into interchromatin granule clusters. J Biol Chem. 2005;280(36):31760–31767. Prepublished on 2005/07/01 as doi:10.1074/jbc.M500884200. - PubMed
-
- Smith MR, Greene WC. Identification of HTLV-I tax trans-activator mutants exhibiting novel transcriptional phenotypes. Genes Dev. 1990;4(11):1875–1885. Prepublished on 1990/11/01 as doi:10.1101/gad.4.11.1875. - PubMed
-
- Viollet B, Lefrancois-Martinez AM, Henrion A, Kahn A, Raymondjean M, Martinez A. Immunochemical characterization and transacting properties of upstream stimulatory factor isoforms. J Biol Chem. 1996;271(3):1405–1415. Prepublished on 1996/01/19 as doi:10.1074/jbc.271.3.1405. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

