Yeast telomere maintenance is globally controlled by programmed ribosomal frameshifting and the nonsense-mediated mRNA decay pathway
- PMID: 24563826
- PMCID: PMC3908577
- DOI: 10.4161/trla.24418
Yeast telomere maintenance is globally controlled by programmed ribosomal frameshifting and the nonsense-mediated mRNA decay pathway
Abstract
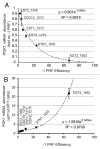
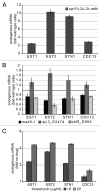
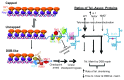
We have previously shown that ~10% of all eukaryotic mRNAs contain potential programmed -1 ribosomal frameshifting (-1 PRF) signals and that some function as mRNA destabilizing elements through the Nonsense-Mediated mRNA Decay (NMD) pathway by directing translating ribosomes to premature termination codons. Here, the connection between -1 PRF, NMD and telomere end maintenance are explored. Functional -1 PRF signals were identified in the mRNAs encoding two components of yeast telomerase, EST1 and EST2, and in mRNAs encoding proteins involved in recruiting telomerase to chromosome ends, STN1 and CDC13. All of these elements responded to mutants and drugs previously known to stimulate or inhibit -1 PRF, further supporting the hypothesis that they promote -1 PRF through the canonical mechanism. All affected the steady-state abundance of a reporter mRNA and the wide range of -1 PRF efficiencies promoted by these elements enabled the determination of an inverse logarithmic relationship between -1 PRF efficiency and mRNA accumulation. Steady-state abundances of the endogenous EST1, EST2, STN1 and CDC13 mRNAs were similarly inversely proportional to changes in -1 PRF efficiency promoted by mutants and drugs, supporting the hypothesis that expression of these genes is post-transcriptionally controlled by -1 PRF under native conditions. Overexpression of EST2 by ablation of -1 PRF signals or inhibition of NMD promoted formation of shorter telomeres and accumulation of large budded cells at the G2/M boundary. A model is presented describing how limitation and maintenance of correct stoichiometries of telomerase components by -1 PRF is used to maintain yeast telomere length.
Keywords: Nonsense-mediated decay; cell cycle; frameshifting; mRNA; ribosome; telomerase.
Figures


References
-
- Dinman JD. . Mechanisms and implications of programmed translational frameshifting. Wiley Interdiscip Rev RNA 2012; 3:661 - 73; http://dx.doi.org/10.1002/wrna.1126; PMID: 22715123 - DOI - PMC - PubMed
-
- Harger JW, Meskauskas A, Dinman JD. . An “integrated model” of programmed ribosomal frameshifting. Trends Biochem Sci 2002; 27:448 - 54; http://dx.doi.org/10.1016/S0968-0004(02)02149-7; PMID: 12217519 - DOI - PubMed
-
- Kontos H, Napthine S, Brierley I. . Ribosomal pausing at a frameshifter RNA pseudoknot is sensitive to reading phase but shows little correlation with frameshift efficiency. Mol Cell Biol 2001; 21:8657 - 70; http://dx.doi.org/10.1128/MCB.21.24.8657-8670.2001; PMID: 11713298 - DOI - PMC - PubMed
-
- Lopinski JD, Dinman JD, Bruenn JA. . Kinetics of ribosomal pausing during programmed -1 translational frameshifting. Mol Cell Biol 2000; 20:1095 - 103; http://dx.doi.org/10.1128/MCB.20.4.1095-1103.2000; PMID: 10648594 - DOI - PMC - PubMed
-
- Plant EP, Jacobs KLM, Harger JW, Meskauskas A, Jacobs JL, Baxter JL, et al. . . The 9-A solution: how mRNA pseudoknots promote efficient programmed -1 ribosomal frameshifting. RNA 2003; 9:168 - 74; http://dx.doi.org/10.1261/rna.2132503; PMID: 12554858 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
