libgapmis: extending short-read alignments
- PMID: 24564250
- PMCID: PMC3821552
- DOI: 10.1186/1471-2105-14-S11-S4
libgapmis: extending short-read alignments
Abstract
Background: A wide variety of short-read alignment programmes have been published recently to tackle the problem of mapping millions of short reads to a reference genome, focusing on different aspects of the procedure such as time and memory efficiency, sensitivity, and accuracy. These tools allow for a small number of mismatches in the alignment; however, their ability to allow for gaps varies greatly, with many performing poorly or not allowing them at all. The seed-and-extend strategy is applied in most short-read alignment programmes. After aligning a substring of the reference sequence against the high-quality prefix of a short read--the seed--an important problem is to find the best possible alignment between a substring of the reference sequence succeeding and the remaining suffix of low quality of the read--extend. The fact that the reads are rather short and that the gap occurrence frequency observed in various studies is rather low suggest that aligning (parts of) those reads with a single gap is in fact desirable.
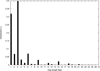
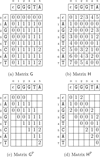
Results: In this article, we present libgapmis, a library for extending pairwise short-read alignments. Apart from the standard CPU version, it includes ultrafast SSE- and GPU-based implementations. libgapmis is based on an algorithm computing a modified version of the traditional dynamic-programming matrix for sequence alignment. Extensive experimental results demonstrate that the functions of the CPU version provided in this library accelerate the computations by a factor of 20 compared to other programmes. The analogous SSE- and GPU-based implementations accelerate the computations by a factor of 6 and 11, respectively, compared to the CPU version. The library also provides the user the flexibility to split the read into fragments, based on the observed gap occurrence frequency and the length of the read, thereby allowing for a variable, but bounded, number of gaps in the alignment.
Conclusions: We present libgapmis, a library for extending pairwise short-read alignments. We show that libgapmis is better-suited and more efficient than existing algorithms for this task. The importance of our contribution is underlined by the fact that the provided functions may be seamlessly integrated into any short-read alignment pipeline. The open-source code of libgapmis is available at http://www.exelixis-lab.org/gapmis.
Figures

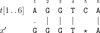





References
-
- Levenshtein VI. Tech Rep 8. Soviet Physics Doklady; 1966. Binary codes capable of correcting deletions, insertions, and reversals.
-
- Wagner RA, Fischer MJ. The String-to-String Correction Problem. Journal of the ACM. 1974;21:168–173. doi: 10.1145/321796.321811. - DOI
-
- Sellers PH. On the Theory and Computation of Evolutionary Distances. SIAM Journal on Applied Mathematics. 1974;26(4):787–793. doi: 10.1137/0126070. - DOI
-
- Heckel P. A technique for isolating differences between files. Communications of the ACM. 1978;21(4):264–268. doi: 10.1145/359460.359467. - DOI
-
- Peterson JL. Computer programs for detecting and correcting spelling errors. Communications of the ACM. 1980;23(12):676–687. doi: 10.1145/359038.359041. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

