MetaSel: a metaphase selection tool using a Gaussian-based classification technique
- PMID: 24564477
- PMCID: PMC4015449
- DOI: 10.1186/1471-2105-14-S16-S13
MetaSel: a metaphase selection tool using a Gaussian-based classification technique
Abstract
Background: Identification of good metaphase spreads is an important step in chromosome analysis for identifying individuals with genetic disorders. The process of finding suitable metaphase chromosomes for accurate clinical analysis is, however, very time consuming since they are selected manually. The selection of suitable metaphase chromosome spreads thus represents a major bottleneck for conventional cytogenetic analysis. Although many algorithms have been developed for karyotyping, none have adequately addressed the critical bottleneck of selecting suitable chromosome spreads. In this paper, we present a software tool that uses a simple rule-based system to efficiently identify metaphase spreads suitable for karyotyping.
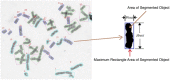
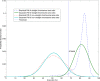
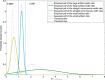
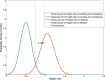
Results: The chromosome shapes can be classified by the software into four main classes. The first and the second classes refer to individual chromosomes with straight and skewed shapes, respectively. The third class is characterized as those chromosomes with overlapping bodies and the fourth class is for the non-chromosome objects. Good metaphase spreads should largely contain chromosomes of the first and the second classes, while the third class should be kept minimal. Several image parameters were examined and used for creating rule-based classification. The threshold value for each parameter is determined using a statistical model. We observed that the Gaussian model can represent the empirical probability density function of the parameters and, hence, the threshold value can be easily determined. The proposed rules can efficiently and accurately classify the individual chromosome with > 90% accuracy.
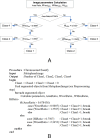
Conclusions: The software tool, termed MetaSel, was developed. Using the Gaussian-based rules, the tool can be used to quickly rank hundreds of chromosome spread images so as to assist cytogeneticists to perform karyotyping effectively. Furthermore, MetaSel offers an intuitive, yet comprehensive, workflow to assist karyotyping, including tools for editing chromosome (split, merge and fix) and a karyotyping editor (moving, rotating, and pairing homologous chromosomes). The program can be freely downloaded from "http://www4a.biotec.or.th/GI/tools/metasel".
Figures






References
-
- Castleman Kenneth R. The PSI Automatic MetaphaseFinder. Journal of Radiation Research. 1992;14(Suppl 1):124–128. - PubMed
-
- Huber R, Kulka U, Lörch T, Braselmann H, Bauchinger M. Automated metaphase finding: an assessment of the efficiency of the METAFER2 system in a routine mutagenicity assay. Mutation Research/ Environmental Mutagenesis and Related Subjects. 1995;14:(1) 97–102. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

