Reconstructing the modular recombination history of Staphylococcus aureus phages
- PMID: 24564731
- PMCID: PMC3852002
- DOI: 10.1186/1471-2105-14-S15-S17
Reconstructing the modular recombination history of Staphylococcus aureus phages
Abstract
Background: Viruses that infect bacteria, called phages, are well-known for their extreme mosaicism, in which an individual genome shares many different parts with many others. The mechanisms for creating these mosaics are largely unknown but are believed to be recombinations, either illegitimate, or partly homologous. In order to reconstruct the history of these recombinations, we need to identify the positions where recombinations may have occurred, and develop algorithms to generate and explore the possible reconstructions.
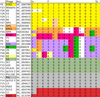
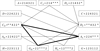
Results: We first show that, provided that their gene order is co-linear, genomes of phages can be aligned, even if large parts of their sequences lack any detectable similarity and are annotated hypothetical proteins. We give such an alignment for 31 Staphylococcus aureus phage genomes, and algorithms that can be used in any similar context. These alignments provide the datasets needed for a combinatorial study of recombinations. We next reconstruct the most likely recombination history of the set of 31 phages, under the hypothesis that recombinations are partly homologous. This history relies on the computational identification of missing phages.
Conclusions: This first combinatorial study of modular recombinations acts as a proof of concept. We show that alignments of whole genomes are feasible for large sets of phages, and that this representation yields data that can be used to reconstruct parts of the evolutionary history of these organisms.
Figures

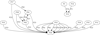
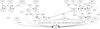
References
-
- Kececioglu JD, Gusfield D. Reconstructing a History of Recombinations From a Set of Sequences. Discrete Applied Mathematics. 1998;14(1-3):239–260. doi: 10.1016/S0166-218X(98)00074-2. - DOI
-
- Ukkonen E. In: WABI'2002, Volume 2452 of Lecture Notes in Computer Science. Guigó R, Gusfield D, editor. Springer; 2002. Finding Founder Sequences from a Set of Recombinants; pp. 277–286.
-
- Wu Y, Gusfield D. In: CPM'2007, Volume 4580 of Lecture Notes in Computer Science. Ma B, Zhang K, editor. Springer; 2007. Improved Algorithms for Inferring the Minimum Mosaic of a Set of Recombinants; pp. 150–161.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

