The impact of structural diversity and parameterization on maps of the protein universe
- PMID: 24565442
- PMCID: PMC4029320
- DOI: 10.1186/1753-6561-7-S7-S1
The impact of structural diversity and parameterization on maps of the protein universe
Abstract
Background: Low dimensional maps of protein structure space (MPSS) provide a powerful global representation of all proteins. In such mappings structural relationships are depicted through spatial adjacency of points, each of which represents a molecule. MPSS can help in understanding the local and global topological characteristics of the structure space, as well as elucidate structure-function relationships within and between sets of proteins. A number of meta- and method-dependent parameters are involved in creating MPSS. However, at the state-of-the-art, a systematic investigation of the influence of these parameters on MPSS construction has yet to be carried out. Further, while specific cases in which MPSS out-perform pairwise distances for prediction of functional annotations have been noted, no general explanation for this phenomenon has yet been advanced.
Methods: We address the above questions within the technical context of creating MPSS by utilizing multidimensional scaling (MDS) for obtaining low-dimensional projections of structure alignment distances.
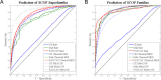
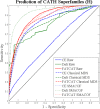
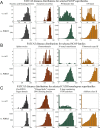
Results and conclusion: MDS is demonstrated as an effective method for construction of MPSS where related structures are co-located, even when their functional and evolutionary proximity cannot be deduced from distributions of pairwise comparisons alone. In particular, we show that MPSS exceed pairwise distance distributions in predictive capability for those annotations of shared function or origin which are characterized by a high level of structural diversity. We also determine the impact of the choice of structure alignment and MDS algorithms on the accuracy of such predictions.
Figures


References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

