The developmental transcriptome of the human brain: implications for neurodevelopmental disorders
- PMID: 24565942
- PMCID: PMC4038354
- DOI: 10.1097/WCO.0000000000000069
The developmental transcriptome of the human brain: implications for neurodevelopmental disorders
Abstract
Purpose of review: Recent characterizations of the transcriptome of the developing human brain by several groups have generated comprehensive datasets on coding and noncoding RNAs that will be instrumental for illuminating the underlying biology of complex neurodevelopmental disorders. This review summarizes recent studies successfully utilizing these data to increase our understanding of the molecular mechanisms of pathogenesis.
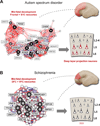
Recent findings: Several approaches have successfully integrated developmental transcriptome data with gene discovery to generate testable hypotheses about when and where in the developing human brain disease-associated genes converge. Specifically, these include the projection neurons in the prefrontal and primary motor--somatosensory cortex during mid-fetal development in autism spectrum disorder and the frontal cortex during fetal development in schizophrenia.
Summary: Developmental transcriptome data is a key to interpreting disease-associated mutations and transcriptional changes. Novel approaches integrating the spatial and temporal dimensions of these data have increased our understanding of when and where disease occurs. Refinement of spatial and temporal properties and expanding these findings to other neurodevelopmental disorders will provide critical insights for understanding disease biology.
Conflict of interest statement
Conflicts of Interest: There are no conflicts of interest
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

