Doripenem, gentamicin, and colistin, alone and in combinations, against gentamicin-susceptible, KPC-producing Klebsiella pneumoniae strains with various ompK36 genotypes
- PMID: 24566172
- PMCID: PMC4068457
- DOI: 10.1128/AAC.01949-13
Doripenem, gentamicin, and colistin, alone and in combinations, against gentamicin-susceptible, KPC-producing Klebsiella pneumoniae strains with various ompK36 genotypes
Abstract
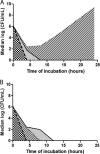
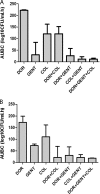
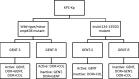
Gentamicin doses of 2 and 10 μg/ml were bactericidal against 64% and 100%, respectively, of gentamicin-susceptible KPC-2-producing Klebsiella pneumoniae strains. Treatment with the combination of doripenem (8 μg/ml) plus colistin (2 μg/ml) was inferior to treatment with gentamicin (2 μg/ml), doripenem-gentamicin, gentamicin-colistin, and doripenem-gentamicin-colistin against strains with glycine and aspartic acid insertions in OpmK36 porin at amino acid (aa) positions 134 and 135 (n = 9). Doripenem-colistin was comparable to other 2- or 3-drug regimens and superior to single drugs against wild-type/minor ompK36 mutants (n = 5). An algorithm incorporating ompK36 genotypes and susceptibility to gentamicin and doripenem may predict antimicrobial activity against KPC-producing K. pneumoniae.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures

References
-
- Qureshi ZA, Paterson DL, Potoski BA, Kilayko MC, Sandovsky G, Sordillo E, Polsky B, Adams-Haduch JM, Doi Y. 2012. Treatment outcome of bacteremia due to KPC-producing Klebsiella pneumoniae: superiority of combination antimicrobial regimens. Antimicrob. Agents Chemother. 56:2108-2113. 10.1128/AAC.06268-11 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

