Origin plasticity during budding yeast DNA replication in vitro
- PMID: 24566988
- PMCID: PMC3989655
- DOI: 10.1002/embj.201387278
Origin plasticity during budding yeast DNA replication in vitro
Abstract
The separation of DNA replication origin licensing and activation in the cell cycle is essential for genome stability across generations in eukaryotic cells. Pre-replicative complexes (pre-RCs) license origins by loading Mcm2-7 complexes in inactive form around DNA. During origin firing in S phase, replisomes assemble around the activated Mcm2-7 DNA helicase. Budding yeast pre-RCs have previously been reconstituted in vitro with purified proteins. Here, we show that reconstituted pre-RCs support replication of plasmid DNA in yeast cell extracts in a reaction that exhibits hallmarks of cellular replication initiation. Plasmid replication in vitro results in the generation of covalently closed circular daughter molecules, indicating that the system recapitulates the initiation, elongation, and termination stages of DNA replication. Unexpectedly, yeast origin DNA is not strictly required for DNA replication in vitro, as heterologous DNA sequences could support replication of plasmid molecules. Our findings support the notion that epigenetic mechanisms are important for determining replication origin sites in budding yeast, highlighting mechanistic principles of replication origin specification that are common among eukaryotes.
Figures

Left: Reaction scheme; blue circle indicates plasmid, gray sphere indicates magnetic bead. Reaction products were analyzed in lanes 1–6 by autoradiography after alkaline agarose gel electrophoresis.
Left: Reaction scheme. Lanes 1–3: Autoradiogram of replication products fractionated by alkaline agarose gel electrophoresis. Lanes 4–6: Western blot analysis of proteins associated with DNA after wash step. Pre-RCs were treated after Dbf4-dependent kinase (DDK) phosphorylation, as indicated on top, prior to transfer into S-phase extract. Low-salt wash: 0.3 M K-glutamate buffer; high-salt wash: 0.5 M NaCl buffer.
Left: Reaction scheme. Lanes 1–10: Western blot analysis of indicated proteins associating with DNA beads after 40-min incubation in S-phase extract. Pre-ICs were assembled in S-phase extract prepared from YDR89 (lanes 1–5) or YSD8 (lanes 6–10) cells.
Autoradiogram of replication products obtained following reaction scheme as in (C). Purified DNA was isolated after the reaction and analyzed by alkaline agarose gel electrophoresis.

Reaction scheme.
Time-course analysis of pARS1 replication. Replication products were analyzed by autoradiography after native (lanes 1–5) or denaturing alkaline (lanes 6–10) agarose gel electrophoresis. Total 32P-dCTP incorporation as determined by phosphorimaging is plotted over time on the right.
Replication in S-phase extract containing 1.25% DMSO (lanes 1, 3) or 177 μM ICRF-193 (lanes 2, 4). Replication products were analyzed by autoradiography after native (lanes 1, 2) or alkaline (lanes 3, 4) agarose gel electrophoresis. Histogram on the left shows total 32P-dCTP incorporation as determined by PhosphorImager analysis; averages and standard deviations of three experimental replicates are shown. HMW: high molecular weight DNA; ssl: single-stranded linear; ccc: covalently closed circular.
Replication reactions were carried out as in (A), DNA isolated from the replication reaction, and either mock-treated (lane 1) or treated with exonuclease III (Exo III, lane 2) prior to native agarose gel electrophoresis.
DNA isolated from a replication reaction was either mock-treated (lanes 1, 4), or treated with 0.4 units (lanes 2, 5) or 2 units (lanes 3, 6) of DpnI, and analyzed by native agarose gel electrophoresis. An ethidium bromide stain of the gel is shown on the left (lanes 1–3), the corresponding autoradiogram is depicted on the right (lanes 4–6).

Time-course analysis of pARS1 replication in the absence (lanes 1–4, and 9–12) or presence (lanes 5–8, and 13–16) of Cdc6 during the pre-RC assembly reaction. Autoradiograms of native (lanes 1–8) or alkaline (lanes 9–16) agarose gel analyses of replication products are shown; total 32P-dCTP incorporation is plotted on the right.
Replication reactions were carried out in the absence or presence of factors indicated on top of each panel. Replication products were analyzed by native (lanes 1–5) or alkaline (lanes 6–10) agarose gel electrophoresis and autoradiography. Total 32P-dCTP incorporation is plotted on the left.
In vitro replication reaction in which purified Cdt1·Mcm2-7 was omitted (lane 1) or included (lane 2) during pre-RC assembly. Replication products were analyzed by native agarose gel electrophoresis. Histogram depicts the total 32P-dCTP incorporation.
32P-dCTP incorporation was monitored in mock-depleted, Mcm10-myc-depleted, or Mcm10-myc-depleted extract supplemented with purified recombinant Mcm10.
In vitro replication was performed in the presence of 1.25% DMSO (lanes 1, 3) or 37 μM aphidicolin (lanes 2, 4). Products were analyzed by native (lanes 1–2) or alkaline (lanes 3–4) agarose gel electrophoresis and autoradiography. Total 32P-dCTP incorporation is plotted on the left.

In vitro replication of wild-type or mutant pARS1 (7 nM) at various origin recognition complex (ORC) concentrations as indicated. Shown is the autoradiogram of replication products separated by native agarose gel electrophoresis.
Total 32P-dCTP incorporation, respectively, as in (A).

Map of pARS1 showing the location of restriction sites used for plasmid fragmentation; resulting restriction fragment sizes are indicated in bold font.
Association of purified ORC with restriction fragments derived from pARS1-wt (lanes 1–20) or pARS1-A−B2− (lanes 21–40). 32P-labeled fragments co-precipitating with ORC were analyzed by native agarose gel electrophoresis and autoradiography. ORC concentrations are indicated on top. Binding was performed in the presence or absence of ATP as indicated. Fragment cartoons are shown on the right.
Mcm2-7 loading on pARS1-wt and pARS1-A−B2− fragments. Fragments co-precipitating with FLAG-Mcm3 in the presence of ATP (lanes 1–2 and 5–6) or ATPγS (lanes 3–4 and 7–8) were analyzed by native agarose gel electrophoresis and autoradiography. Immunoprecipitates were washed with 0.5 M NaCl-containing buffer (high-salt wash) instead of 0.3 M K-glutamate-containing buffer as indicated.

DNase1 footprint analysis of purified origin recognition complex (ORC) bound to a CEN4-containing fragment. “A-rich” denotes the A-rich strand, “T-rich” denotes the T-rich strand of the ACS in CDE III. Orange boxes indicate the region of DNase1 protection by ORC, arrowheads indicate ORC-induced DNase1 hypersensitive sites. The positions of CEN4 sequence elements are indicated by blue boxes.
Sequence comparison of the ORC binding sites at CEN4 and ARS1. Orange boxes indicate regions of ORC protection from DNase1 cleavage; ACS matches are highlighted in red underlined font; blue boxes indicate CEN4 elements; gray boxes indicate the positions of the ARS1 A and B1 elements.
Total 32P-dCTP incorporation as measured by phosphorimaging after in vitro replication of pARS1-A−B2−/CDE III-ACS− or pARS1-A−B2− in the presence of 0, 2, 7, and 27 nM ORC. Shown are the averages of a duplicate experiment. Error bars are too small to be depicted.
Mcm2-7 loading on pARS1-A−B2−/CDE III-A− or pARS1-A−B2− restriction fragments as measured by co-immunoprecipitation of radiolabelled restriction fragments with FLAG-MCM3.
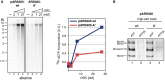
Replication of pARS305-wt and pARS305-A− (7 nM, respectively) at limiting origin recognition complex (ORC) concentrations in vitro. Replication products were analyzed by autoradiography after denaturing alkaline agarose gel electrophoresis (lanes 1–8). Graph shows total 32P-dCTP incorporation, respectively, as determined from duplicate experiments. Error bars are too small to be depicted.
Reconstituted pre-RCs were assembled using 10 nM ORC in the presence of ATP or ATPγS on bead-coupled wild-type and mutant pARS305, as indicated, washed with 0.5 M NaCl buffer (high-salt wash), and analyzed by SDS–PAGE and silver stain.

Map of pARS305. Orange and blue boxes demarcate the genomic fragment integrated into pUC19.
Association of purified ORC with restriction fragments derived from pARS305-wt (lanes 1–20) or pARS305-A− (lanes 21–40), analogous to Fig 5B.
Mcm2-7 loading on pARS305-wt and pARS305-A− fragments as in Fig 5C. Total DNA recovered by IP, as determined by PhosphorImager analysis, is indicated on top in percent of the amount recovered in lane 1.

Reconstituted pre-RC assembly in the presence of ATP or ATPγS on bead-coupled pBluescript, washed with 0.5 M NaCl buffer (high-salt wash) as indicated, and analyzed by SDS–PAGE and silver stain.
In vitro replication of pBluescript. Reactions were performed in the presence of indicated concentrations of origin recognition complex (ORC) and replication products analyzed by autoradiography after native agarose gel electrophoresis.
Comparison of in vitro replication efficiencies of pBluescript and pARS1. Input plasmid concentration was 7 nM. Relative 32P-dCTP incorporation values were normalized for G/C-content and respective plasmid length.
Reconstituted pre-RC assembly on pET16b and pARS1 (ORC: 10 nM). All reactions were subjected to high-salt wash after assembly and analyzed by SDS–PAGE and silver stain.
In vitro replication of pARS1 and pET16b. Replication products were analyzed by native agarose gel electrophoresis and autoradiography.
Quantitation of total 32P-dCTP incorporation in experiment in (E).
Relative in vitro replication activity of pET16b, pET16b containing a 254 bp autonomously replicating sequence (ARS) 1 fragment (pET16b/ARS1), and pET16b containing the 1.1-kb CEN4-containing fragment of pARS1 (see Fig 5A; pET16b/CEN4 fragment). Averages and standard deviations of three independent experiments are shown.
References
-
- Araki H. Cyclin-dependent kinase-dependent initiation of chromosomal DNA replication. Curr Opin Cell Biol. 2010;22:766–771. - PubMed
-
- Beall EL, Manak JR, Zhou S, Bell M, Lipsick JS, Botchan MR. Role for a Drosophila Myb-containing protein complex in site-specific DNA replication. Nature. 2002;420:833–837. - PubMed
-
- Bell SP, Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992;357:128–134. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

