Origin of β-carotene-rich plastoglobuli in Dunaliella bardawil
- PMID: 24567188
- PMCID: PMC3982768
- DOI: 10.1104/pp.113.235119
Origin of β-carotene-rich plastoglobuli in Dunaliella bardawil
Abstract
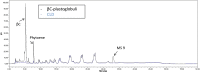
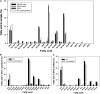
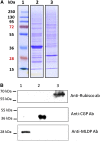
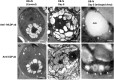
The halotolerant microalgae Dunaliella bardawil accumulates under nitrogen deprivation two types of lipid droplets: plastoglobuli rich in β-carotene (βC-plastoglobuli) and cytoplasmatic lipid droplets (CLDs). We describe the isolation, composition, and origin of these lipid droplets. Plastoglobuli contain β-carotene, phytoene, and galactolipids missing in CLDs. The two preparations contain different lipid-associated proteins: major lipid droplet protein in CLD and the Prorich carotene globule protein in βC-plastoglobuli. The compositions of triglyceride (TAG) molecular species, total fatty acids, and sn-1+3 and sn-2 positions in the two lipid pools are similar, except for a small increase in palmitic acid in plastoglobuli, suggesting a common origin. The formation of CLD TAG precedes that of βC-plastoglobuli, reaching a maximum after 48 h of nitrogen deprivation and then decreasing. Palmitic acid incorporation kinetics indicated that, at early stages of nitrogen deprivation, CLD TAG is synthesized mostly from newly formed fatty acids, whereas in βC-plastoglobuli, a large part of TAG is produced from fatty acids of preformed membrane lipids. Electron microscopic analyses revealed that CLDs adhere to chloroplast envelope membranes concomitant with appearance of small βC-plastoglobuli within the chloroplast. Based on these results, we propose that CLDs in D. bardawil are produced in the endoplasmatic reticulum, whereas βC-plastoglobuli are made, in part, from hydrolysis of chloroplast membrane lipids and in part, by a continual transfer of TAG or fatty acids derived from CLD.
Figures







Similar articles
-
Proteome analysis of cytoplasmatic and plastidic β-carotene lipid droplets in Dunaliella bardawil.Plant Physiol. 2015 Jan;167(1):60-79. doi: 10.1104/pp.114.248450. Epub 2014 Nov 17. Plant Physiol. 2015. PMID: 25404729 Free PMC article.
-
Novel 9-cis/all-trans β-carotene isomerases from plastidic oil bodies in Dunaliella bardawil catalyze the conversion of all-trans to 9-cis β-carotene.Plant Cell Rep. 2017 Jun;36(6):807-814. doi: 10.1007/s00299-017-2110-7. Epub 2017 Mar 11. Plant Cell Rep. 2017. PMID: 28285407
-
A hypothesis about the origin of carotenoid lipid droplets in the green algae Dunaliella and Haematococcus.Planta. 2019 Jan;249(1):31-47. doi: 10.1007/s00425-018-3050-3. Epub 2018 Nov 23. Planta. 2019. PMID: 30470898 Review.
-
Effect of dissolved inorganic carbon on β-carotene and fatty acid production in Dunaliella sp.Appl Biochem Biotechnol. 2015 Mar;175(6):2895-906. doi: 10.1007/s12010-014-1461-6. Epub 2015 Jan 10. Appl Biochem Biotechnol. 2015. PMID: 25575588
-
Metabolic and functional connections between cytoplasmic and chloroplast triacylglycerol storage.Prog Lipid Res. 2020 Nov;80:101069. doi: 10.1016/j.plipres.2020.101069. Epub 2020 Oct 27. Prog Lipid Res. 2020. PMID: 33127353 Review.
Cited by
-
Enhanced acetyl-CoA production is associated with increased triglyceride accumulation in the green alga Chlorella desiccata.J Exp Bot. 2015 Jul;66(13):3725-35. doi: 10.1093/jxb/erv166. Epub 2015 Apr 28. J Exp Bot. 2015. PMID: 25922486 Free PMC article.
-
The cadmium-induced changes in the polar and neutral lipid compositions suggest the involvement of triacylglycerol in the defense response in maize.Physiol Mol Biol Plants. 2020 Jan;26(1):15-23. doi: 10.1007/s12298-019-00734-9. Epub 2019 Dec 9. Physiol Mol Biol Plants. 2020. PMID: 32158117 Free PMC article.
-
Cultivation modes affect the morphology, biochemical composition, and antioxidant and anti-inflammatory properties of the green microalga Neochloris oleoabundans.Protoplasma. 2024 Nov;261(6):1185-1206. doi: 10.1007/s00709-024-01958-7. Epub 2024 Jun 12. Protoplasma. 2024. PMID: 38864933 Free PMC article.
-
Proteome analysis of cytoplasmatic and plastidic β-carotene lipid droplets in Dunaliella bardawil.Plant Physiol. 2015 Jan;167(1):60-79. doi: 10.1104/pp.114.248450. Epub 2014 Nov 17. Plant Physiol. 2015. PMID: 25404729 Free PMC article.
-
Analysis of triglyceride synthesis unveils a green algal soluble diacylglycerol acyltransferase and provides clues to potential enzymatic components of the chloroplast pathway.BMC Genomics. 2017 Mar 9;18(1):223. doi: 10.1186/s12864-017-3602-0. BMC Genomics. 2017. PMID: 28274201 Free PMC article.
References
-
- Al-Hasan RH, Ghannoum MA, Sallal AK, Abu-Elteen KH, Radwan SS. (1987) Correlative changes of growth, pigmentation and lipid composition of Dunaliella salina in response to halostress. J Gen Microbiol 133: 2607–2616
-
- Avron A, Ben-Amotz A. (1992) Dunaliella: Physiology, Biochemistry and Biotechnology. CRC Press, Boca Raton, FL
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

