The myc 3' wnt-responsive element suppresses colonic tumorigenesis
- PMID: 24567369
- PMCID: PMC3993608
- DOI: 10.1128/MCB.00969-13
The myc 3' wnt-responsive element suppresses colonic tumorigenesis
Abstract
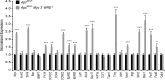
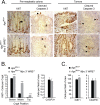
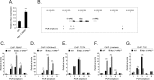
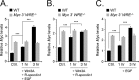
Mutations in components of the Wnt/β-catenin signaling pathway are commonly found in colorectal cancers, and these mutations cause aberrant expression of genes controlled by Wnt-responsive DNA elements (WREs). While the c-Myc proto-oncogene (Myc) is required for intestinal phenotypes associated with pathogenic Wnt/β-catenin signaling in vivo, the WREs that control Myc expression in this setting have yet to be fully described. Previously, we demonstrated that the Myc 3' WRE was required for intestinal homeostasis and intestinal repair in response to damage. Here, we tested the role of the Myc 3' WRE in intestinal tumorigenesis using two independent mouse models. In comparison to Apc(Min/+) mice, Apc(Min/+) Myc 3' WRE(-/-) mice contained 25% fewer tumors in the small intestine. Deletion of the Myc 3' WRE(-/-) in the Apc(Min/+) background resulted in 4-fold more colonic tumors. In a model of colitis-associated colorectal cancer, the Myc 3' WRE suppressed colonic tumorigenesis, most notably within the cecum. Using chromatin immunoprecipitation and transcript analysis of purified colonic crypts, we found that the Myc 3' WRE is required for the transcriptional regulation of Myc expression in vivo. These results emphasize the critical role of the Myc 3' WRE in maintaining homeostatic Myc expression.
Figures




Similar articles
-
Regulation of MYC gene expression by aberrant Wnt/β-catenin signaling in colorectal cancer.World J Biol Chem. 2015 Nov 26;6(4):290-300. doi: 10.4331/wjbc.v6.i4.290. World J Biol Chem. 2015. PMID: 26629312 Free PMC article. Review.
-
A dynamic exchange of TCF3 and TCF4 transcription factors controls MYC expression in colorectal cancer cells.Cell Cycle. 2015;14(3):323-32. doi: 10.4161/15384101.2014.980643. Cell Cycle. 2015. PMID: 25659031 Free PMC article.
-
A hypermorphic epithelial β-catenin mutation facilitates intestinal tumorigenesis in mice in response to compounding WNT-pathway mutations.Dis Model Mech. 2015 Nov;8(11):1361-73. doi: 10.1242/dmm.019844. Epub 2015 Aug 6. Dis Model Mech. 2015. PMID: 26398937 Free PMC article.
-
The Myc 3' Wnt responsive element regulates neutrophil recruitment after acute colonic injury in mice.Dig Dis Sci. 2013 Oct;58(10):2858-2867. doi: 10.1007/s10620-013-2686-x. Epub 2013 May 3. Dig Dis Sci. 2013. PMID: 23640071 Free PMC article.
-
Transcriptional Regulation of Wnt/β-Catenin Pathway in Colorectal Cancer.Cells. 2020 Sep 19;9(9):2125. doi: 10.3390/cells9092125. Cells. 2020. PMID: 32961708 Free PMC article. Review.
Cited by
-
A Role for MYC in Lithium-Stimulated Repair of the Colonic Epithelium After DSS-Induced Damage in Mice.Dig Dis Sci. 2016 Feb;61(2):410-22. doi: 10.1007/s10620-015-3852-0. Epub 2015 Aug 30. Dig Dis Sci. 2016. PMID: 26320084
-
Regenerating Reputations: Are Wnt and Myc the Good Guys After All?Dig Dis Sci. 2016 Feb;61(2):327-9. doi: 10.1007/s10620-015-3947-7. Dig Dis Sci. 2016. PMID: 26520110 No abstract available.
-
Regulation of MYC gene expression by aberrant Wnt/β-catenin signaling in colorectal cancer.World J Biol Chem. 2015 Nov 26;6(4):290-300. doi: 10.4331/wjbc.v6.i4.290. World J Biol Chem. 2015. PMID: 26629312 Free PMC article. Review.
-
Increased Musashi-2 and Decreased NUMB Protein Levels Observed in Human Colorectal Cancer are reverted to Normal Levels by ATRA-Induced Cell Differentiation.Int J Cancer Res Ther. 2018;3(2):10.33140/ijcrt/03/02/00003. doi: 10.33140/ijcrt/03/02/00003. Epub 2018 Aug 15. Int J Cancer Res Ther. 2018. PMID: 32984754 Free PMC article.
-
Tristetraprolin targets Nos2 expression in the colonic epithelium.Sci Rep. 2019 Oct 8;9(1):14413. doi: 10.1038/s41598-019-50957-9. Sci Rep. 2019. PMID: 31595002 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
