A comprehensive, high-resolution map of a gene's fitness landscape
- PMID: 24567513
- PMCID: PMC4032126
- DOI: 10.1093/molbev/msu081
A comprehensive, high-resolution map of a gene's fitness landscape
Erratum in
-
A Comprehensive, High-Resolution Map of a Gene's Fitness Landscape.Mol Biol Evol. 2016 May;33(5):1378. doi: 10.1093/molbev/msw021. Epub 2016 Feb 23. Mol Biol Evol. 2016. PMID: 26912810 Free PMC article. No abstract available.
Abstract
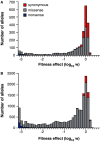
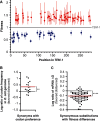
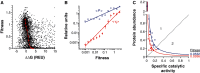
Mutations are central to evolution, providing the genetic variation upon which selection acts. A mutation's effect on the suitability of a gene to perform a particular function (gene fitness) can be positive, negative, or neutral. Knowledge of the distribution of fitness effects (DFE) of mutations is fundamental for understanding evolutionary dynamics, molecular-level genetic variation, complex genetic disease, the accumulation of deleterious mutations, and the molecular clock. We present comprehensive DFEs for point and codon mutants of the Escherichia coli TEM-1 β-lactamase gene and missense mutations in the TEM-1 protein. These DFEs provide insight into the inherent benefits of the genetic code's architecture, support for the hypothesis that mRNA stability dictates codon usage at the beginning of genes, an extensive framework for understanding protein mutational tolerance, and evidence that mutational effects on protein thermodynamic stability shape the DFE. Contrary to prevailing expectations, we find that deleterious effects of mutation primarily arise from a decrease in specific protein activity and not cellular protein levels.
Keywords: beta-lactamase; fitness landscape; protein evolution.
© The Author 2014. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

