Cytoplasmic 5'-3' exonuclease Xrn1p is also a genome-wide transcription factor in yeast
- PMID: 24567736
- PMCID: PMC3915102
- DOI: 10.3389/fgene.2014.00001
Cytoplasmic 5'-3' exonuclease Xrn1p is also a genome-wide transcription factor in yeast
Abstract
The 5' to 3' exoribonuclease Xrn1 is a large protein involved in cytoplasmatic mRNA degradation as a critical component of the major decaysome. Its deletion in the yeast Saccharomyces cerevisiae is not lethal, but it has multiple physiological effects. In a previous study, our group showed that deletion of all tested components of the yeast major decaysome, including XRN1, results in a decrease in the synthetic rate and an increase in half-life of most mRNAs in a compensatory manner. Furthermore, the same study showed that the all tested decaysome components are also nuclear proteins that bind to the 5' region of a number of genes. In the present work, we show that disruption of Xrn1 activity preferentially affects both the synthesis and decay of a distinct subpopulation of mRNAs. The most affected mRNAs are the transcripts of the highly transcribed genes, mainly those encoding ribosome biogenesis and translation factors. Previously, we proposed that synthegradases play a key role in regulating both mRNA synthesis and degradation. Evidently, Xrn1 functions as a synthegradase, whose selectivity might help coordinating the expression of the protein synthetic machinery. We propose to name the most affected genes "Xrn1 synthegradon."
Keywords: mRNA decay; mRNA stability; mRNA synthesis; nascent transcription; transcription rate.
Figures
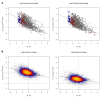

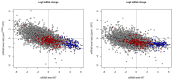
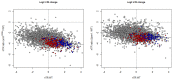


References
-
- Alberola T. M., García-Martínez J., Antúnez O., Viladevall L., Barceló A., Ariño J., et al. (2004). A new set of DNA macrochips for the yeast Saccharomyces cerevisiae: features and uses. Int. Microbiol. 7, 199–206 - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

