Acetyl-lysine binding site of bromodomain-containing protein 4 (BRD4) interacts with diverse kinase inhibitors
- PMID: 24568369
- PMCID: PMC4032195
- DOI: 10.1021/cb500072z
Acetyl-lysine binding site of bromodomain-containing protein 4 (BRD4) interacts with diverse kinase inhibitors
Abstract
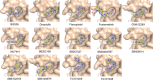
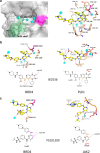
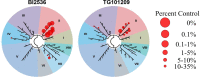
Members of the bromodomain and extra terminal (BET) family of proteins are essential for the recognition of acetylated lysine (KAc) residues in histones and have emerged as promising drug targets in cancer, inflammation, and contraception research. In co-crystallization screening campaigns using the first bromodomain of BRD4 (BRD4-1) against kinase inhibitor libraries, we identified and characterized 14 kinase inhibitors (10 distinct chemical scaffolds) as ligands of the KAc binding site. Among these, the PLK1 inhibitor BI2536 and the JAK2 inhibitor TG101209 displayed strongest inhibitory potential against BRD4 (IC50=25 nM and 130 nM, respectively) and high selectivity for BET bromodomains. Comparative structural analysis revealed markedly different binding modes of kinase hinge-binding scaffolds in the KAc binding site, suggesting that BET proteins are potential off-targets of diverse kinase inhibitors. Combined, these findings provide a new structural framework for the rational design of next-generation BET-selective and dual-activity BET-kinase inhibitors.
Figures


Similar articles
-
Covalent-Fragment Screening of BRD4 Identifies a Ligandable Site Orthogonal to the Acetyl-Lysine Binding Sites.ACS Chem Biol. 2020 Apr 17;15(4):1036-1049. doi: 10.1021/acschembio.0c00058. Epub 2020 Mar 23. ACS Chem Biol. 2020. PMID: 32149490 Free PMC article.
-
Structural and Atropisomeric Factors Governing the Selectivity of Pyrimido-benzodiazipinones as Inhibitors of Kinases and Bromodomains.ACS Chem Biol. 2018 Sep 21;13(9):2438-2448. doi: 10.1021/acschembio.7b00638. Epub 2018 Aug 31. ACS Chem Biol. 2018. PMID: 30102854 Free PMC article.
-
Metabolically Derived Lysine Acylations and Neighboring Modifications Tune the Binding of the BET Bromodomains to Histone H4.Biochemistry. 2017 Oct 17;56(41):5485-5495. doi: 10.1021/acs.biochem.7b00595. Epub 2017 Oct 5. Biochemistry. 2017. PMID: 28945351 Free PMC article.
-
Progress in the development and application of small molecule inhibitors of bromodomain-acetyl-lysine interactions.J Med Chem. 2012 Nov 26;55(22):9393-413. doi: 10.1021/jm300915b. Epub 2012 Sep 27. J Med Chem. 2012. PMID: 22924434 Review.
-
BET Bromodomain as a Target of Epigenetic Therapy.Chem Pharm Bull (Tokyo). 2016;64(6):540-7. doi: 10.1248/cpb.c16-00225. Chem Pharm Bull (Tokyo). 2016. PMID: 27250788 Review.
Cited by
-
Pharmacological Targeting of BET Bromodomain Proteins in Acute Myeloid Leukemia and Malignant Lymphomas: From Molecular Characterization to Clinical Applications.Cancers (Basel). 2019 Oct 2;11(10):1483. doi: 10.3390/cancers11101483. Cancers (Basel). 2019. PMID: 31581671 Free PMC article. Review.
-
A Structure-based Design Approach for Generating High Affinity BRD4 D1-Selective Chemical Probes.J Med Chem. 2022 Feb 10;65(3):2342-2360. doi: 10.1021/acs.jmedchem.1c01779. Epub 2022 Jan 10. J Med Chem. 2022. PMID: 35007061 Free PMC article.
-
The bromodomain: from epigenome reader to druggable target.Biochim Biophys Acta. 2014 Aug;1839(8):676-85. doi: 10.1016/j.bbagrm.2014.03.011. Epub 2014 Mar 28. Biochim Biophys Acta. 2014. PMID: 24686119 Free PMC article. Review.
-
Selective chemical modulation of gene transcription favors oligodendrocyte lineage progression.Chem Biol. 2014 Jul 17;21(7):841-854. doi: 10.1016/j.chembiol.2014.05.009. Epub 2014 Jun 19. Chem Biol. 2014. PMID: 24954007 Free PMC article.
-
Targeting bromodomains: epigenetic readers of lysine acetylation.Nat Rev Drug Discov. 2014 May;13(5):337-56. doi: 10.1038/nrd4286. Epub 2014 Apr 22. Nat Rev Drug Discov. 2014. PMID: 24751816 Review.
References
-
- Filippakopoulos P.; Picaud S.; Mangos M.; Keates T.; Lambert J. P.; Barsyte-Lovejoy D.; Felletar I.; Volkmer R.; Muller S.; Pawson T.; Gingras A. C.; Arrowsmith C. H.; Knapp S. (2012) Histone recognition and large-scale structural analysis of the human bromodomain family. Cell 149, 214–231. - PMC - PubMed
-
- Filippakopoulos P.; Qi J.; Picaud S.; Shen Y.; Smith W. B.; Fedorov O.; Morse E. M.; Keates T.; Hickman T. T.; Felletar I.; Philpott M.; Munro S.; McKeown M. R.; Wang Y.; Christie A. L.; West N.; Cameron M. J.; Schwartz B.; Heightman T. D.; La Thangue N.; French C. A.; Wiest O.; Kung A. L.; Knapp S.; Bradner J. E. (2010) Selective inhibition of BET bromodomains. Nature 468, 1067–1073. - PMC - PubMed
-
- French C. A.; Miyoshi I.; Kubonishi I.; Grier H. E.; Perez-Atayde A. R.; Fletcher J. A. (2003) BRD4-NUT fusion oncogene: a novel mechanism in aggressive carcinoma. Cancer Res. 63, 304–307. - PubMed
-
- Darnell J. E. (2002) Transcription factors as targets for cancer therapy. Nat. Rev. Cancer 2, 740–749. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

