Functional genomics reveals that Clostridium difficile Spo0A coordinates sporulation, virulence and metabolism
- PMID: 24568651
- PMCID: PMC4028888
- DOI: 10.1186/1471-2164-15-160
Functional genomics reveals that Clostridium difficile Spo0A coordinates sporulation, virulence and metabolism
Abstract
Background: Clostridium difficile is an anaerobic, Gram-positive bacterium that can reside as a commensal within the intestinal microbiota of healthy individuals or cause life-threatening antibiotic-associated diarrhea in immunocompromised hosts. C. difficile can also form highly resistant spores that are excreted facilitating host-to-host transmission. The C. difficile spo0A gene encodes a highly conserved transcriptional regulator of sporulation that is required for relapsing disease and transmission in mice.
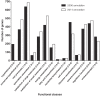
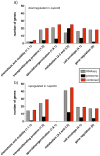
Results: Here we describe a genome-wide approach using a combined transcriptomic and proteomic analysis to identify Spo0A regulated genes. Our results validate Spo0A as a positive regulator of putative and novel sporulation genes as well as components of the mature spore proteome. We also show that Spo0A regulates a number of virulence-associated factors such as flagella and metabolic pathways including glucose fermentation leading to butyrate production.
Conclusions: The C. difficile spo0A gene is a global transcriptional regulator that controls diverse sporulation, virulence and metabolic phenotypes coordinating pathogen adaptation to a wide range of host interactions. Additionally, the rich breadth of functional data allowed us to significantly update the annotation of the C. difficile 630 reference genome which will facilitate basic and applied research on this emerging pathogen.
Figures



References
-
- Walker AS, Eyre DW, Wyllie DH, Dingle KE, Harding RM, O’Connor L, Griffiths D, Vaughan A, Finney J, Wilcox MH, Crook DW, Peto TE. Characterisation of Clostridium difficile hospital ward-based transmission using extensive epidemiological data and molecular typing. PLoS Med. 2012;9(2):e1001172. doi: 10.1371/journal.pmed.1001172. - DOI - PMC - PubMed
-
- He M, Miyajima F, Roberts P, Ellison L, Pickard DJ, Martin MJ, Connor TR, Harris SR, Fairley D, Bamford KB, D'Arc S, Brazier J, Brown D, Coia JE, Douce G, Gerding D, Kim HJ, Koh TH, Kato H, Senoh M, Louie T, Michell S, Butt E, Peacock SJ, Brown NM, Riley T, Songer G, Wilcox M, Pirmohamed M, Kuijper E. et al.Emergence and global spread of epidemic healthcare-associated Clostridium difficile. Nat Genet. 2012;45(1):109–113. doi: 10.1038/ng.2478. - DOI - PMC - PubMed
-
- He M, Sebaihia M, Lawley TD, Stabler RA, Dawson LF, Martin MJ, Holt KE, Seth-Smith HM, Quail MA, Rance R, Brooks K, Churcher C, Harris D, Bentley SD, Burrows C, Clark L, Corton C, Murray V, Rose G, Thurston S, van Tonder A, Walker D, Wren BW, Dougan G, Parkhill J. Evolutionary dynamics of Clostridium difficile over short and long time scales. Proc Natl Acad Sci USA. 2010;107(16):7527–7532. doi: 10.1073/pnas.0914322107. - DOI - PMC - PubMed
-
- Sebaihia M, Wren BW, Mullany P, Fairweather NF, Minton N, Stabler R, Thomson NR, Roberts AP, Cerdeno-Tarraga AM, Wang H, Holden MT, Wright A, Churcher C, Quail MA, Baker S, Bason N, Brooks K, Chillingworth T, Cronin A, Davis P, Dowd L, Fraser A, Feltwell T, Hance Z, Holroyd S, Jagels K, Moule S, Mungall K, Price C, Rabbinowitsch E. et al.The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat Genet. 2006;38(7):779–786. doi: 10.1038/ng1830. - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

