Pegylated IFN-α regulates hepatic gene expression through transient Jak/STAT activation
- PMID: 24569457
- PMCID: PMC3973080
- DOI: 10.1172/JCI70408
Pegylated IFN-α regulates hepatic gene expression through transient Jak/STAT activation
Abstract
The use of pegylated interferon-α (pegIFN-α) has replaced unmodified recombinant IFN-α for the treatment of chronic viral hepatitis. While the superior antiviral efficacy of pegIFN-α is generally attributed to improved pharmacokinetic properties, the pharmacodynamic effects of pegIFN-α in the liver have not been studied. Here, we analyzed pegIFN-α-induced signaling and gene regulation in paired liver biopsies obtained prior to treatment and during the first week following pegIFN-α injection in 18 patients with chronic hepatitis C. Despite sustained high concentrations of pegIFN-α in serum, the Jak/STAT pathway was activated in hepatocytes only on the first day after pegIFN-α administration. Evaluation of liver biopsies revealed that pegIFN-α induces hundreds of genes that can be classified into four clusters based on different temporal expression profiles. In all clusters, gene transcription was mainly driven by IFN-stimulated gene factor 3 (ISGF3). Compared with conventional IFN-α therapy, pegIFN-α induced a broader spectrum of gene expression, including many genes involved in cellular immunity. IFN-induced secondary transcription factors did not result in additional waves of gene expression. Our data indicate that the superior antiviral efficacy of pegIFN-α is not the result of prolonged Jak/STAT pathway activation in hepatocytes, but rather is due to induction of additional genes that are involved in cellular immune responses.
Figures


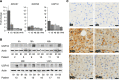



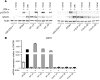

Comment in
-
New insight into the enhanced effect of pegylated interferon-α.Hepatology. 2014 Oct;60(4):1435-7. doi: 10.1002/hep.27269. Epub 2014 Aug 25. Hepatology. 2014. PMID: 24944103 No abstract available.
References
-
- Larner AC, Chaudhuri A, Darnell JE, Darnell JE., Jr Transcriptional induction by interferon. New protein(s) determine the extent and length of the induction. J Biol Chem. 1986;261(1):453–459. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

