Structural basis for the prion-like MAVS filaments in antiviral innate immunity
- PMID: 24569476
- PMCID: PMC3932521
- DOI: 10.7554/eLife.01489
Structural basis for the prion-like MAVS filaments in antiviral innate immunity
Erratum in
-
Correction: Structural basis for the prion-like MAVS filaments in antiviral innate immunity.Elife. 2015 Aug 28;4:e07546. doi: 10.7554/eLife.07546. Elife. 2015. PMID: 26314863 Free PMC article. No abstract available.
Abstract
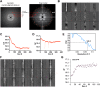
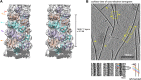
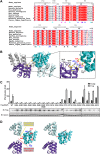
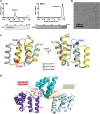
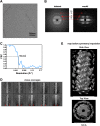
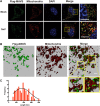
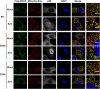
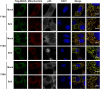
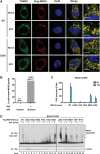
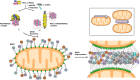
Mitochondrial antiviral signaling (MAVS) protein is required for innate immune responses against RNA viruses. In virus-infected cells MAVS forms prion-like aggregates to activate antiviral signaling cascades, but the underlying structural mechanism is unknown. Here we report cryo-electron microscopic structures of the helical filaments formed by both the N-terminal caspase activation and recruitment domain (CARD) of MAVS and a truncated MAVS lacking part of the proline-rich region and the C-terminal transmembrane domain. Both structures are left-handed three-stranded helical filaments, revealing specific interfaces between individual CARD subunits that are dictated by electrostatic interactions between neighboring strands and hydrophobic interactions within each strand. Point mutations at multiple locations of these two interfaces impaired filament formation and antiviral signaling. Super-resolution imaging of virus-infected cells revealed rod-shaped MAVS clusters on mitochondria. These results elucidate the structural mechanism of MAVS polymerization, and explain how an α-helical domain uses distinct chemical interactions to form self-perpetuating filaments. DOI: http://dx.doi.org/10.7554/eLife.01489.001.
Keywords: MAVS; cryoEM reconstruction; innate immunity; prion-like filaments; three-stranded filaments.
Conflict of interest statement
ZJC: Reviewing editor,
The other authors declare that no competing interests exist.
Figures



Comment in
-
Three-stranded antiviral attack.Elife. 2014 Jan 1;3:e02369. doi: 10.7554/eLife.02369. Elife. 2014. PMID: 24596154 Free PMC article.
References
-
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. 2002. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallographica Section D Biological Crystallography 58:1948–1954. 10.1107/S0907444902016657 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

