Molybdenum metabolism in plants and crosstalk to iron
- PMID: 24570679
- PMCID: PMC3916724
- DOI: 10.3389/fpls.2014.00028
Molybdenum metabolism in plants and crosstalk to iron
Abstract
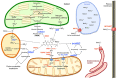
In the form of molybdate the transition metal molybdenum is essential for plants as it is required by a number of enzymes that catalyze key reactions in nitrogen assimilation, purine degradation, phytohormone synthesis, and sulfite detoxification. However, molybdate itself is biologically inactive and needs to be complexed by a specific organic pterin in order to serve as a permanently bound prosthetic group, the molybdenum cofactor, for the socalled molybdo-enyzmes. While the synthesis of molybdenum cofactor has been intensively studied, only little is known about the uptake of molybdate by the roots, its transport to the shoot and its allocation and storage within the cell. Yet, recent evidence indicates that intracellular molybdate levels are tightly controlled by molybdate transporters, in particular during plant development. Moreover, a tight connection between molybdenum and iron metabolisms is presumed because (i) uptake mechanisms for molybdate and iron affect each other, (ii) most molybdo-enzymes do also require iron-containing redox groups such as iron-sulfur clusters or heme, (iii) molybdenum metabolism has recruited mechanisms typical for iron-sulfur cluster synthesis, and (iv) both molybdenum cofactor synthesis and extramitochondrial iron-sulfur proteins involve the function of a specific mitochondrial ABC-type transporter.
Keywords: aldehyde oxidase; iron; molybdate transporter; molybdenum; molybdo-enzymes; nitrate reductase; sulfite oxidase; xanthine dehydrogenase.
Figures
References
-
- Akaba S., Seo M., Dohmae N., Takio K., Sekimoto H., Kamiya Y., et al. (1999). Production of homo- and hetero-dimeric isozymes from two aldehyde oxidase genes of Arabidopsis thaliana. J. Biochem. 126 395–401 - PubMed
-
- Baxter I., Muthukumar B., Park H. C., Buchner P., Lahner B., Danku J., et al. (2008). Variation in molybdenum content across broadly distributed populations of Arabidopsis thaliana is controlled by a mitochondrial molybdenum transporter (MOT1). PLoS Genet. 4:e1000004 10.1371/journal.pgen.1000004 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

