In situ visualization of newly synthesized proteins in environmental microbes using amino acid tagging and click chemistry
- PMID: 24571640
- PMCID: PMC4122687
- DOI: 10.1111/1462-2920.12436
In situ visualization of newly synthesized proteins in environmental microbes using amino acid tagging and click chemistry
Abstract
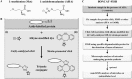
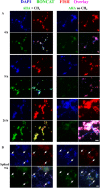
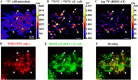
Here we describe the application of a new click chemistry method for fluorescent tracking of protein synthesis in individual microorganisms within environmental samples. This technique, termed bioorthogonal non-canonical amino acid tagging (BONCAT), is based on the in vivo incorporation of the non-canonical amino acid L-azidohomoalanine (AHA), a surrogate for l-methionine, followed by fluorescent labelling of AHA-containing cellular proteins by azide-alkyne click chemistry. BONCAT was evaluated with a range of phylogenetically and physiologically diverse archaeal and bacterial pure cultures and enrichments, and used to visualize translationally active cells within complex environmental samples including an oral biofilm, freshwater and anoxic sediment. We also developed combined assays that couple BONCAT with ribosomal RNA (rRNA)-targeted fluorescence in situ hybridization (FISH), enabling a direct link between taxonomic identity and translational activity. Using a methanotrophic enrichment culture incubated under different conditions, we demonstrate the potential of BONCAT-FISH to study microbial physiology in situ. A direct comparison of anabolic activity using BONCAT and stable isotope labelling by nano-scale secondary ion mass spectrometry ((15)NH(3) assimilation) for individual cells within a sediment-sourced enrichment culture showed concordance between AHA-positive cells and (15)N enrichment. BONCAT-FISH offers a fast, inexpensive and straightforward fluorescence microscopy method for studying the in situ activity of environmental microbes on a single-cell level.
© 2014 The Authors. Environmental Microbiology published by Society for Applied Microbiology and John Wiley & Sons Ltd.
Figures




References
-
- Adachi T, Yamada Y. Inoue I. Alternative method for selective reduction of unsaturated nucleoside azides to amines. Synthesis-Stuttgart. 1977;1977:45–46.
-
- Agard NJ, Prescher JA. Bertozzi CR. A strain-promoted [3 + 2] azide-alkyne cycloaddition for covalent modification of biomolecules in living systems. J Am Chem Soc. 2004;126:15046–15047. - PubMed
-
- Bagert JD, Xie YJ, Sweredoski MJ, Qi Y, Hess S, Schuman EM. Tirrell DA. Quantitative, time-resolved proteomic analysis by combining bioorthogonal noncanonical amino acid tagging and pulsed stable isotope labeling by amino acids in cell culture. Mol Cell Proteomics. 2014 doi: 10.1074/mcp.M113.031914 mcp.M113.031914. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

