Genome-wide association mapping of acute lung injury in neonatal inbred mice
- PMID: 24571919
- PMCID: PMC4021442
- DOI: 10.1096/fj.13-247221
Genome-wide association mapping of acute lung injury in neonatal inbred mice
Abstract
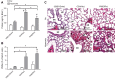
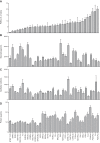
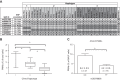
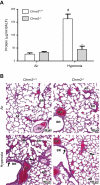
Reactive oxygen species (ROS) contribute to the pathogenesis of many acute and chronic pulmonary disorders, including bronchopulmonary dysplasia (BPD), a respiratory condition that affects preterm infants. However, the mechanisms of susceptibility to oxidant stress in neonatal lungs are not completely understood. We evaluated the role of genetic background in response to oxidant stress in the neonatal lung by exposing mice from 36 inbred strains to hyperoxia (95% O2) for 72 h after birth. Hyperoxia-induced lung injury was evaluated by using bronchoalveolar lavage fluid (BALF) analysis and pathology. Statistically significant interstrain variation was found for BALF inflammatory cells and protein (heritability estimates range: 33.6-55.7%). Genome-wide association mapping using injury phenotypes identified quantitative trait loci (QTLs) on chromosomes 1, 2, 4, 6, and 7. Comparative mapping of the chromosome 6 QTLs identified Chrm2 (cholinergic receptor, muscarinic 2, cardiac) as a candidate susceptibility gene, and mouse strains with a nonsynonymous coding single-nucleotide polymorphism (SNP) in Chrm2 that causes an amino acid substitution (P265L) had significantly reduced hyperoxia-induced inflammation compared to strains without the SNP. Further, hyperoxia-induced lung injury was significantly reduced in neonatal mice with targeted deletion of Chrm2, relative to wild-type controls. This study has important implications for understanding the mechanisms of oxidative lung injury in neonates.
Keywords: bronchopulmonary dysplasia; cardiac; cholinergic receptor; inflammation; muscarinic 2; quantitative trait locus.
© FASEB.
Figures

References
-
- Northway W. H., Jr., Rosan R. C., Porter D. Y. (1967) Pulmonary disease following respirator therapy of hyaline-membrane disease: bronchopulmonary dysplasia. N. Engl. J. Med. 276, 357–368 - PubMed
-
- Jobe A. H., Bancalari E. (2001) Bronchopulmonary dysplasia. Am. J. Respir. Crit. Care Med. 163, 1723–1729 - PubMed
-
- Bhandari A., Bhandari V. (2009) Pitfalls, problems, and progress in bronchopulmonary dysplasia. Pediatrics 123, 1562–1573 - PubMed
-
- Parton L. A., Strassberg S. S., Qian D., Galvin-Parton P. A., Cristea I. A. (2006) The genetic basis for bronchopulmonary dysplasia. Front. Biosci. 11, 1854–1860 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

