Probing bunyavirus N protein oligomerisation using mass spectrometry
- PMID: 24573811
- PMCID: PMC4377080
- DOI: 10.1002/rcm.6841
Probing bunyavirus N protein oligomerisation using mass spectrometry
Abstract
Rationale: Bunyaviruses have become a major threat to both humans and livestock in Europe and the Americas. The nucleocapsid (N) protein of these viruses is key to the replication cycle and knowledge of the N oligomerisation state is central to understanding the viral lifecycle and for development of therapeutic strategies.
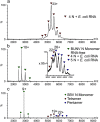
Methods: Bunyamwera virus and Schmallenberg virus N proteins (BUNV-N and SBV-N) were expressed recombinantly in E. coli as hexahistidine-SUMO-tagged fusions, and the tag removed subsequently. Noncovalent nano-electrospray ionisation mass spectrometry was conducted in the presence and absence of short RNA oligonucleotides. Instrumental conditions were optimised for the transmission of intact protein complexes into the gas phase. The resulting protein-protein and protein-RNA complexes were identified and their stoichiometries verified by their mass. Collision-induced dissociation tandem mass spectrometry was used in cases of ambiguity.
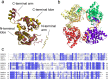
Results: Both BUNV-N and SBV-N proteins reassembled into N-RNA complexes in the presence of RNA; however, SBV-N formed a wider range of complexes with varying oligomeric states. The N:RNA oligomers observed were consistent with a model of assembly via stepwise addition of N proteins. Furthermore, upon mixing the two proteins in the presence of RNA no heteromeric complexes were observed, thus revealing insights into the specificity of oligomerisation.
Conclusions: Noncovalent mass spectrometry has provided the first detailed analysis of the co-populated oligomeric species formed by these important viral proteins and revealed insights into their assembly pathways. Using this technique has also enabled comparisons to be made between the two N proteins.
Copyright © 2014 John Wiley & Sons, Ltd.
Figures


Similar articles
-
Nucleocapsid protein structures from orthobunyaviruses reveal insight into ribonucleoprotein architecture and RNA polymerization.Nucleic Acids Res. 2013 Jun;41(11):5912-26. doi: 10.1093/nar/gkt268. Epub 2013 Apr 17. Nucleic Acids Res. 2013. PMID: 23595147 Free PMC article.
-
Investigating the specificity and stoichiometry of RNA binding by the nucleocapsid protein of Bunyamwera virus.RNA. 2009 Mar;15(3):391-9. doi: 10.1261/rna.1367209. Epub 2009 Jan 23. RNA. 2009. PMID: 19168749 Free PMC article.
-
Expression and purification of the nucleocapsid protein of Schmallenberg virus, and preparation and characterization of a monoclonal antibody against this protein.Protein Expr Purif. 2013 Nov;92(1):1-8. doi: 10.1016/j.pep.2013.08.012. Epub 2013 Aug 26. Protein Expr Purif. 2013. PMID: 23988909
-
The nucleocapsid of vesicular stomatitis virus.Sci China Life Sci. 2012 Apr;55(4):291-300. doi: 10.1007/s11427-012-4307-x. Epub 2012 May 9. Sci China Life Sci. 2012. PMID: 22566085 Review.
-
Nucleic-acid-chaperone activity of retroviral nucleocapsid proteins: significance for viral replication.Trends Biochem Sci. 1998 Aug;23(8):297-301. doi: 10.1016/s0968-0004(98)01256-0. Trends Biochem Sci. 1998. PMID: 9757830 Review.
Cited by
-
The Native Orthobunyavirus Ribonucleoprotein Possesses a Helical Architecture.mBio. 2022 Aug 30;13(4):e0140522. doi: 10.1128/mbio.01405-22. Epub 2022 Jun 28. mBio. 2022. PMID: 35762594 Free PMC article.
-
Asymmetric Trimeric Ring Structure of the Nucleocapsid Protein of Tospovirus.J Virol. 2017 Sep 27;91(20):e01002-17. doi: 10.1128/JVI.01002-17. Print 2017 Oct 15. J Virol. 2017. PMID: 28768868 Free PMC article.
-
Chemical cross-linking and native mass spectrometry: A fruitful combination for structural biology.Protein Sci. 2015 Aug;24(8):1193-209. doi: 10.1002/pro.2696. Epub 2015 May 27. Protein Sci. 2015. PMID: 25970732 Free PMC article. Review.
References
-
- Maltezou HC, Papa A. Crimean-Congo hemorrhagic fever: risk for emergence of new endemic foci in Europe? Travel Med. Infect. Dis. 2010;8:139. - PubMed
-
- Dunn EF, Pritlove DC, Jin H, Elliott RM. Transcription of a recombinant RNA bunyavirus template by transiently expressed bunyavirus proteins. Virology. 1995;211:133. - PubMed
-
- Elliott RM. Nucleotide sequence analysis of the large (L) genomic RNA segment of Bunyamwera virus, the prototype of the family Bunyaviridae. Virology. 1989;173:426. - PubMed
-
- Elliott RM. Identification of nonstructural proteins encoded by viruses of the Bunyamwera serogroup (family Bunyaviridae) Virology. 1985;143:119. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
