COMPARTMENTS: unification and visualization of protein subcellular localization evidence
- PMID: 24573882
- PMCID: PMC3935310
- DOI: 10.1093/database/bau012
COMPARTMENTS: unification and visualization of protein subcellular localization evidence
Abstract
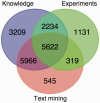
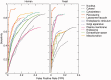
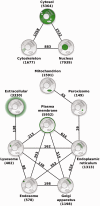
Information on protein subcellular localization is important to understand the cellular functions of proteins. Currently, such information is manually curated from the literature, obtained from high-throughput microscopy-based screens and predicted from primary sequence. To get a comprehensive view of the localization of a protein, it is thus necessary to consult multiple databases and prediction tools. To address this, we present the COMPARTMENTS resource, which integrates all sources listed above as well as the results of automatic text mining. The resource is automatically kept up to date with source databases, and all localization evidence is mapped onto common protein identifiers and Gene Ontology terms. We further assign confidence scores to the localization evidence to facilitate comparison of different types and sources of evidence. To further improve the comparability, we assign confidence scores based on the type and source of the localization evidence. Finally, we visualize the unified localization evidence for a protein on a schematic cell to provide a simple overview. Database URL: http://compartments.jensenlab.org.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

