Hox proteins drive cell segregation and non-autonomous apical remodelling during hindbrain segmentation
- PMID: 24574009
- PMCID: PMC3957373
- DOI: 10.1242/dev.098954
Hox proteins drive cell segregation and non-autonomous apical remodelling during hindbrain segmentation
Abstract
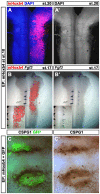
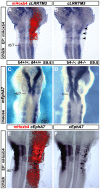
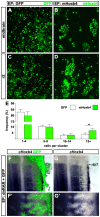
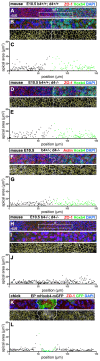
Hox genes encode a conserved family of homeodomain transcription factors regulating development along the major body axis. During embryogenesis, Hox proteins are expressed in segment-specific patterns and control numerous different segment-specific cell fates. It has been unclear, however, whether Hox proteins drive the epithelial cell segregation mechanism that is thought to initiate the segmentation process. Here, we investigate the role of vertebrate Hox proteins during the partitioning of the developing hindbrain into lineage-restricted units called rhombomeres. Loss-of-function mutants and ectopic expression assays reveal that Hoxb4 and its paralogue Hoxd4 are necessary and sufficient for cell segregation, and for the most caudal rhombomere boundary (r6/r7). Hox4 proteins regulate Eph/ephrins and other cell-surface proteins, and can function in a non-cell-autonomous manner to induce apical cell enlargement on both sides of their expression border. Similarly, other Hox proteins expressed at more rostral rhombomere interfaces can also regulate Eph/ephrins, induce apical remodelling and drive cell segregation in ectopic expression assays. However, Krox20, a key segmentation factor expressed in odd rhombomeres (r3 and r5), can largely override Hox proteins at the level of regulation of a cell surface target, Epha4. This study suggests that most, if not all, Hox proteins share a common potential to induce cell segregation but in some contexts this is masked or modulated by other transcription factors.
Keywords: Apical polarity; Cell affinities; Cell segregation; Cell sorting; Cell tension; Chick; Hindbrain; Hox; Mouse; Rhombomeres; Segmentation.
Figures



Similar articles
-
Rostral hindbrain patterning involves the direct activation of a Krox20 transcriptional enhancer by Hox/Pbx and Meis factors.Development. 2008 Oct;135(20):3369-78. doi: 10.1242/dev.023614. Epub 2008 Sep 11. Development. 2008. PMID: 18787068
-
Rhombomere transplantation repatterns the segmental organization of cranial nerves and reveals cell-autonomous expression of a homeodomain protein.Development. 1993 Jan;117(1):105-17. doi: 10.1242/dev.117.1.105. Development. 1993. PMID: 7900983
-
Paralog group 1 hox genes regulate rhombomere 5/6 expression of vhnf1, a repressor of rostral hindbrain fates, in a meis-dependent manner.Dev Biol. 2004 Jul 15;271(2):350-61. doi: 10.1016/j.ydbio.2004.04.007. Dev Biol. 2004. PMID: 15223339
-
Hindbrain induction and patterning during early vertebrate development.Cell Mol Life Sci. 2019 Mar;76(5):941-960. doi: 10.1007/s00018-018-2974-x. Epub 2018 Dec 5. Cell Mol Life Sci. 2019. PMID: 30519881 Free PMC article. Review.
-
Developing roles for Hox proteins in hindbrain gene regulatory networks.Int J Dev Biol. 2018;62(11-12):767-774. doi: 10.1387/ijdb.180141cs. Int J Dev Biol. 2018. PMID: 30604846 Review.
Cited by
-
A single-cell transcriptional timelapse of mouse embryonic development, from gastrula to pup.bioRxiv [Preprint]. 2023 Apr 5:2023.04.05.535726. doi: 10.1101/2023.04.05.535726. bioRxiv. 2023. Update in: Nature. 2024 Feb;626(8001):1084-1093. doi: 10.1038/s41586-024-07069-w. PMID: 37066300 Free PMC article. Updated. Preprint.
-
A Simple Predictive Enhancer Syntax for Hindbrain Patterning Is Conserved in Vertebrate Genomes.PLoS One. 2015 Jul 1;10(7):e0130413. doi: 10.1371/journal.pone.0130413. eCollection 2015. PLoS One. 2015. PMID: 26131856 Free PMC article.
-
Hox genes in development and beyond.Development. 2023 Jan 1;150(1):dev192476. doi: 10.1242/dev.192476. Epub 2023 Jan 16. Development. 2023. PMID: 36645372 Free PMC article.
-
An atlas of anterior hox gene expression in the embryonic sea lamprey head: Hox-code evolution in vertebrates.Dev Biol. 2019 Sep 1;453(1):19-33. doi: 10.1016/j.ydbio.2019.05.001. Epub 2019 May 6. Dev Biol. 2019. PMID: 31071313 Free PMC article.
-
Regulating Retinoic Acid Availability during Development and Regeneration: The Role of the CYP26 Enzymes.J Dev Biol. 2020 Mar 5;8(1):6. doi: 10.3390/jdb8010006. J Dev Biol. 2020. PMID: 32151018 Free PMC article. Review.
References
-
- Alexander T., Nolte C., Krumlauf R. (2009). Hox genes and segmentation of the hindbrain and axial skeleton. Annu. Rev. Cell Dev. Biol. 25, 431–456 - PubMed
-
- Araujo M., Nieto M. A. (1997). The expression of chick EphA7 during segmentation of the central and peripheral nervous system. Mech. Dev. 68, 173–177 - PubMed
-
- Barrow J. R., Stadler H. S., Capecchi M. R. (2000). Roles of Hoxa1 and Hoxa2 in patterning the early hindbrain of the mouse. Development 127, 933–944 - PubMed
-
- Becker N., Seitanidou T., Murphy P., Mattéi M. G., Topilko P., Nieto M. A., Wilkinson D. G., Charnay P., Gilardi-Hebenstreit P. (1994). Several receptor tyrosine kinase genes of the Eph family are segmentally expressed in the developing hindbrain. Mech. Dev. 47, 3–17 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

