Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli
- PMID: 24574290
- PMCID: PMC3993690
- DOI: 10.1128/JCM.03617-13
Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli
Abstract
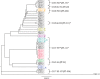
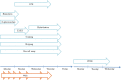
Fast and accurate identification and typing of pathogens are essential for effective surveillance and outbreak detection. The current routine procedure is based on a variety of techniques, making the procedure laborious, time-consuming, and expensive. With whole-genome sequencing (WGS) becoming cheaper, it has huge potential in both diagnostics and routine surveillance. The aim of this study was to perform a real-time evaluation of WGS for routine typing and surveillance of verocytotoxin-producing Escherichia coli (VTEC). In Denmark, the Statens Serum Institut (SSI) routinely receives all suspected VTEC isolates. During a 7-week period in the fall of 2012, all incoming isolates were concurrently subjected to WGS using IonTorrent PGM. Real-time bioinformatics analysis was performed using web-tools (www.genomicepidemiology.org) for species determination, multilocus sequence type (MLST) typing, and determination of phylogenetic relationship, and a specific VirulenceFinder for detection of E. coli virulence genes was developed as part of this study. In total, 46 suspected VTEC isolates were characterized in parallel during the study. VirulenceFinder proved successful in detecting virulence genes included in routine typing, explicitly verocytotoxin 1 (vtx1), verocytotoxin 2 (vtx2), and intimin (eae), and also detected additional virulence genes. VirulenceFinder is also a robust method for assigning verocytotoxin (vtx) subtypes. A real-time clustering of isolates in agreement with the epidemiology was established from WGS, enabling discrimination between sporadic and outbreak isolates. Overall, WGS typing produced results faster and at a lower cost than the current routine. Therefore, WGS typing is a superior alternative to conventional typing strategies. This approach may also be applied to typing and surveillance of other pathogens.
Figures

Similar articles
-
Evaluation of whole-genome sequencing for outbreak detection of Verotoxigenic Escherichia coli O157:H7 from the Canadian perspective.BMC Genomics. 2018 Dec 4;19(1):870. doi: 10.1186/s12864-018-5243-3. BMC Genomics. 2018. PMID: 30514209 Free PMC article.
-
Prevalence of Escherichia coli O157 and verocytotoxin producing E. coli (VTEC) on Danish beef carcasses.Int J Food Microbiol. 2010 Jun 30;141(1-2):90-6. doi: 10.1016/j.ijfoodmicro.2010.03.009. Epub 2010 Mar 25. Int J Food Microbiol. 2010. PMID: 20427097
-
Detection and characterization of verocytotoxin-producing Escherichia coli by automated 5' nuclease PCR assay.J Clin Microbiol. 2003 Jul;41(7):2884-93. doi: 10.1128/JCM.41.7.2884-2893.2003. J Clin Microbiol. 2003. PMID: 12843017 Free PMC article.
-
Characteristics of the Shiga-toxin-producing enteroaggregative Escherichia coli O104:H4 German outbreak strain and of STEC strains isolated in Spain.Int Microbiol. 2011 Sep;14(3):121-41. doi: 10.2436/20.1501.01.142. Int Microbiol. 2011. PMID: 22101411 Review.
-
Bacterial genome sequencing in clinical microbiology: a pathogen-oriented review.Eur J Clin Microbiol Infect Dis. 2017 Nov;36(11):2007-2020. doi: 10.1007/s10096-017-3024-6. Epub 2017 Jun 21. Eur J Clin Microbiol Infect Dis. 2017. PMID: 28639162 Free PMC article. Review.
Cited by
-
Rapid Proteomic Characterization of Bacteriocin-Producing Enterococcus faecium Strains from Foodstuffs.Int J Mol Sci. 2022 Nov 10;23(22):13830. doi: 10.3390/ijms232213830. Int J Mol Sci. 2022. PMID: 36430310 Free PMC article.
-
Genomic Characterization of Two Shiga Toxin-Converting Bacteriophages Induced From Environmental Shiga Toxin-Producing Escherichia coli.Front Microbiol. 2021 Feb 25;12:587696. doi: 10.3389/fmicb.2021.587696. eCollection 2021. Front Microbiol. 2021. PMID: 33716997 Free PMC article.
-
Integrated genome-based probiotic relevance and safety evaluation of Lactobacillus reuteri PNW1.PLoS One. 2020 Jul 20;15(7):e0235873. doi: 10.1371/journal.pone.0235873. eCollection 2020. PLoS One. 2020. PMID: 32687505 Free PMC article.
-
Predictors of blaCTX-M-15 in varieties of Escherichia coli genotypes from humans in community settings in Mwanza, Tanzania.BMC Infect Dis. 2016 Apr 29;16:187. doi: 10.1186/s12879-016-1527-x. BMC Infect Dis. 2016. PMID: 27129719 Free PMC article.
-
Detection, Characterization, and Typing of Shiga Toxin-Producing Escherichia coli.Front Microbiol. 2016 Apr 12;7:478. doi: 10.3389/fmicb.2016.00478. eCollection 2016. Front Microbiol. 2016. PMID: 27148176 Free PMC article. Review.
References
-
- Ribot EM, Fair MA, Gautom R, Cameron DN, Hunter SB, Swaminathan B, Barrett TJ. 2006. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog. Dis. 3:59–67. 10.1089/fpd.2006.3.59 - DOI - PubMed
-
- Kauffmann F. 1975. Classification of bacteria. A realistic scheme with special reference to the classification of Salmonella -and Escherichia- species. Munksgaard, Copenhagen, Denmark
-
- Mellmann A, Harmsen D, Cummings CA, Zentz EB, Leopold SR, Rico A, Prior K, Szczepanowski R, Ji Y, Zhang W, McLaughlin SF, Henkhaus JK, Leopold B, Bielaszewska M, Prager R, Brzoska PM, Moore RL, Guenther S, Rothberg JM, Karch H. 2011. Prospective genomic characterization of the German enterohemorrhagic Escherichia coli O104:H4 outbreak by rapid next generation sequencing technology. PLoS One 6:e22751. 10.1371/journal.pone.0022751 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

