Defective interfering influenza virus RNAs: time to reevaluate their clinical potential as broad-spectrum antivirals?
- PMID: 24574404
- PMCID: PMC4019098
- DOI: 10.1128/JVI.03193-13
Defective interfering influenza virus RNAs: time to reevaluate their clinical potential as broad-spectrum antivirals?
Abstract
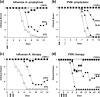
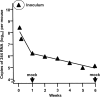
Defective interfering (DI) RNAs are highly deleted forms of the infectious genome that are made by most families of RNA viruses. DI RNAs retain replication and packaging signals, are synthesized preferentially over infectious genomes, and are packaged as DI virus particles which can be transmitted to susceptible cells. Their ability to interfere with the replication of infectious virus in cell culture and their potential as antivirals in the clinic have long been known. However, until now, no realistic formulation has been described. In this review, we consider the early evidence of antiviral activity by DI viruses and, using the example of DI influenza A virus, outline developments that have led to the production of a cloned DI RNA that is highly active in preclinical studies not only against different subtypes of influenza A virus but also against heterologous respiratory viruses. These data suggest the timeliness of reassessing the potential of DI viruses as a novel class of antivirals that may have general applicability.
Figures


Similar articles
-
Generation of a purely clonal defective interfering influenza virus.Microbiol Immunol. 2019 May;63(5):164-171. doi: 10.1111/1348-0421.12681. Epub 2019 May 17. Microbiol Immunol. 2019. PMID: 30997933
-
Cloned Defective Interfering Influenza RNA and a Possible Pan-Specific Treatment of Respiratory Virus Diseases.Viruses. 2015 Jul 8;7(7):3768-88. doi: 10.3390/v7072796. Viruses. 2015. PMID: 26184282 Free PMC article. Review.
-
A Defective Interfering Influenza RNA Inhibits Infectious Influenza Virus Replication in Human Respiratory Tract Cells: A Potential New Human Antiviral.Viruses. 2016 Aug 22;8(8):237. doi: 10.3390/v8080237. Viruses. 2016. PMID: 27556481 Free PMC article.
-
Heterologous protection of mice from a lethal human H1N1 influenza A virus infection by H3N8 equine defective interfering virus: comparison of defective RNA sequences isolated from the DI inoculum and mouse lung.Virology. 1998 Sep 1;248(2):241-53. doi: 10.1006/viro.1998.9267. Virology. 1998. PMID: 9721233
-
Defective interfering viruses and their potential as antiviral agents.Rev Med Virol. 2010 Jan;20(1):51-62. doi: 10.1002/rmv.641. Rev Med Virol. 2010. PMID: 20041441 Review.
Cited by
-
Roles for Pathogen Interference in Influenza Vaccination, with Implications to Vaccine Effectiveness (VE) and Attribution of Influenza Deaths.Infect Dis Rep. 2022 Sep 23;14(5):710-758. doi: 10.3390/idr14050076. Infect Dis Rep. 2022. PMID: 36286197 Free PMC article. Review.
-
Emerging Concepts and Technologies in Vaccine Development.Front Immunol. 2020 Sep 30;11:583077. doi: 10.3389/fimmu.2020.583077. eCollection 2020. Front Immunol. 2020. PMID: 33101309 Free PMC article. Review.
-
Evidence for Viral Interference and Cross-reactive Protective Immunity Between Influenza B Virus Lineages.J Infect Dis. 2018 Jan 30;217(4):548-559. doi: 10.1093/infdis/jix509. J Infect Dis. 2018. PMID: 29325138 Free PMC article.
-
Live Attenuated Influenza Vaccine contains Substantial and Unexpected Amounts of Defective Viral Genomic RNA.Viruses. 2017 Sep 21;9(10):269. doi: 10.3390/v9100269. Viruses. 2017. PMID: 28934167 Free PMC article.
-
Defective viral genomes: critical danger signals of viral infections.J Virol. 2014 Aug;88(16):8720-3. doi: 10.1128/JVI.00707-14. Epub 2014 May 28. J Virol. 2014. PMID: 24872580 Free PMC article. Review.
References
-
- Hurt AC, Chotpitayasunondh T, Cox NJ, Daniels R, Fry AM, Gubareva LV, Hayden FG, Hui SD, Hungnes O, Lackenby A, Lim W, Meijer A, Penn C, Tashiro M, Uyeki TM, Zambon M, WHO Consultation on Pandemic Influenza A (H1N1) 2009 Virus Resistance to Antivirals 2012. Antiviral resistance during the 2009 influenza A H1N1 pandemic: public health, laboratory, and clinical perspectives. Lancet Infect. Dis. 12:240–248. 10.1016/S1473-3099(11)70318-8 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

