Accumulation of aberrant DNA methylation during colorectal cancer development
- PMID: 24574770
- PMCID: PMC3921549
- DOI: 10.3748/wjg.v20.i4.978
Accumulation of aberrant DNA methylation during colorectal cancer development
Abstract
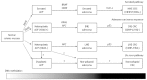
Despite the recent advances in the therapeutic modalities, colorectal cancer (CRC) remains to be one of the most common causes of cancer-related death. CRC arises through accumulation of multiple genetic and epigenetic alterations that transform normal colonic epithelium into adenocarcinomas. Among crucial roles of epigenetic alterations, gene silencing by aberrant DNA methylation of promoter regions is one of the most important epigenetic mechanisms. Recent comprehensive methylation analyses on genome-wide scale revealed that sporadic CRC can be classified into distinct epigenotypes. Each epigenotype cooperates with specific genetic alterations, suggesting that they represent different molecular carcinogenic pathways. Precursor lesions of CRC, such as conventional and serrated adenomas, already show similar methylation accumulation to CRC, and can therefore be classified into those epigenotypes of CRC. In addition, specific DNA methylation already occurs in the normal colonic mucosa, which might be utilized for prediction of the personal CRC risk. DNA methylation is suggested to occur at an earlier stage than carcinoma formation, and may predict the molecular basis for future development of CRC. Here, we review DNA methylation and CRC classification, and discuss the possible clinical usefulness of DNA methylation as biomarkers for the diagnosis, prediction of the prognosis and the response to therapy of CRC.
Keywords: Aberrant crypt foci; Colorectal adenoma; Colorectal cancer; Colorectal carcinogenesis; DNA methylation; Epigenotype; Genetic mutation.
Figures
References
-
- Kaneda A, Feinberg AP. Loss of imprinting of IGF2: a common epigenetic modifier of intestinal tumor risk. Cancer Res. 2005;65:11236–11240. - PubMed
-
- Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, Nakamura Y, White R, Smits AM, Bos JL. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319:525–532. - PubMed
-
- Jen J, Kim H, Piantadosi S, Liu ZF, Levitt RC, Sistonen P, Kinzler KW, Vogelstein B, Hamilton SR. Allelic loss of chromosome 18q and prognosis in colorectal cancer. N Engl J Med. 1994;331:213–221. - PubMed
-
- White RL. Tumor suppressing pathways. Cell. 1998;92:591–592. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

