VH Replacement Footprint Analyzer-I, a Java-Based Computer Program for Analyses of Immunoglobulin Heavy Chain Genes and Potential VH Replacement Products in Human and Mouse
- PMID: 24575092
- PMCID: PMC3918983
- DOI: 10.3389/fimmu.2014.00040
VH Replacement Footprint Analyzer-I, a Java-Based Computer Program for Analyses of Immunoglobulin Heavy Chain Genes and Potential VH Replacement Products in Human and Mouse
Abstract
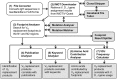
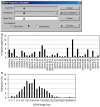
VH replacement occurs through RAG-mediated secondary recombination between a rearranged VH gene and an upstream unrearranged VH gene. Due to the location of the cryptic recombination signal sequence (cRSS, TACTGTG) at the 3' end of VH gene coding region, a short stretch of nucleotides from the previous rearranged VH gene can be retained in the newly formed VH-DH junction as a "footprint" of VH replacement. Such footprints can be used as markers to identify Ig heavy chain (IgH) genes potentially generated through VH replacement. To explore the contribution of VH replacement products to the antibody repertoire, we developed a Java-based computer program, VH replacement footprint analyzer-I (VHRFA-I), to analyze published or newly obtained IgH genes from human or mouse. The VHRFA-1 program has multiple functional modules: it first uses service provided by the IMGT/V-QUEST program to assign potential VH, DH, and JH germline genes; then, it searches for VH replacement footprint motifs within the VH-DH junction (N1) regions of IgH gene sequences to identify potential VH replacement products; it can also analyze the frequencies of VH replacement products in correlation with publications, keywords, or VH, DH, and JH gene usages, and mutation status; it can further analyze the amino acid usages encoded by the identified VH replacement footprints. In summary, this program provides a useful computation tool for exploring the biological significance of VH replacement products in human and mouse.
Keywords: B cell; IGH sequencing; IgH gene; RAG; VDJ rearrangement; VH replacement.
Figures








Similar articles
-
Accumulation of VH Replacement Products in IgH Genes Derived from Autoimmune Diseases and Anti-Viral Responses in Human.Front Immunol. 2014 Jul 22;5:345. doi: 10.3389/fimmu.2014.00345. eCollection 2014. Front Immunol. 2014. PMID: 25101087 Free PMC article.
-
Contribution of V(H) replacement products in mouse antibody repertoire.PLoS One. 2013;8(2):e57877. doi: 10.1371/journal.pone.0057877. Epub 2013 Feb 28. PLoS One. 2013. PMID: 23469094 Free PMC article.
-
Potential contribution of VH gene replacement in immunity and disease.Ann N Y Acad Sci. 2005 Dec;1062:175-81. doi: 10.1196/annals.1358.020. Ann N Y Acad Sci. 2005. PMID: 16461800 Review.
-
VH replacement in primary immunoglobulin repertoire diversification.Proc Natl Acad Sci U S A. 2015 Feb 3;112(5):E458-66. doi: 10.1073/pnas.1418001112. Epub 2015 Jan 21. Proc Natl Acad Sci U S A. 2015. PMID: 25609670 Free PMC article.
-
The molecular basis and biological significance of VH replacement.Immunol Rev. 2004 Feb;197:231-42. doi: 10.1111/j.0105-2896.2004.0107.x. Immunol Rev. 2004. PMID: 14962199 Review.
Cited by
-
Immunologic Basis for Long HCDR3s in Broadly Neutralizing Antibodies Against HIV-1.Front Immunol. 2014 Jun 2;5:250. doi: 10.3389/fimmu.2014.00250. eCollection 2014. Front Immunol. 2014. PMID: 24917864 Free PMC article. Review.
-
Accumulation of VH Replacement Products in IgH Genes Derived from Autoimmune Diseases and Anti-Viral Responses in Human.Front Immunol. 2014 Jul 22;5:345. doi: 10.3389/fimmu.2014.00345. eCollection 2014. Front Immunol. 2014. PMID: 25101087 Free PMC article.
-
Uncommon structural motifs dominate the antigen binding site in human autoantibodies reactive with basement membrane collagen.Mol Immunol. 2016 Aug;76:123-33. doi: 10.1016/j.molimm.2016.07.004. Epub 2016 Jul 20. Mol Immunol. 2016. PMID: 27450516 Free PMC article.
-
Circulating Plasmablasts from Chronically Human Immunodeficiency Virus-Infected Individuals Predominantly Produce Polyreactive/Autoreactive Antibodies.Front Immunol. 2017 Dec 6;8:1691. doi: 10.3389/fimmu.2017.01691. eCollection 2017. Front Immunol. 2017. PMID: 29270169 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

