Reliable differentiation of Meyerozyma guilliermondii from Meyerozyma caribbica by internal transcribed spacer restriction fingerprinting
- PMID: 24575831
- PMCID: PMC3946169
- DOI: 10.1186/1471-2180-14-52
Reliable differentiation of Meyerozyma guilliermondii from Meyerozyma caribbica by internal transcribed spacer restriction fingerprinting
Abstract
Background: Meyerozyma guilliermondii (anamorph Candida guilliermondii) and Meyerozyma caribbica (anamorph Candida fermentati) are closely related species of the genetically heterogenous M. guilliermondii complex. Conventional phenotypic methods frequently misidentify the species within this complex and also with other species of the Saccharomycotina CTG clade. Even the long-established sequencing of large subunit (LSU) rRNA gene remains ambiguous. We also faced similar problem during identification of yeast isolates of M. guilliermondii complex from indigenous bamboo shoot fermentation in North East India. There is a need for development of reliable and accurate identification methods for these closely related species because of their increasing importance as emerging infectious yeasts and associated biotechnological attributes.
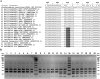
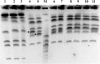
Results: We targeted the highly variable internal transcribed spacer (ITS) region (ITS1-5.8S-ITS2) and identified seven restriction enzymes through in silico analysis for differentiating M. guilliermondii from M. caribbica. Fifty five isolates of M. guilliermondii complex which could not be delineated into species-specific taxonomic ranks by API 20 C AUX and LSU rRNA gene D1/D2 sequencing were subjected to ITS-restriction fragment length polymorphism (ITS-RFLP) analysis. TaqI ITS-RFLP distinctly differentiated the isolates into M. guilliermondii (47 isolates) and M. caribbica (08 isolates) with reproducible species-specific patterns similar to the in silico prediction. The reliability of this method was validated by ITS1-5.8S-ITS2 sequencing, mitochondrial DNA RFLP and electrophoretic karyotyping.
Conclusions: We herein described a reliable ITS-RFLP method for distinct differentiation of frequently misidentified M. guilliermondii from M. caribbica. Even though in silico analysis differentiated other closely related species of M. guilliermondii complex from the above two species, it is yet to be confirmed by in vitro analysis using reference strains. This method can be used as a reliable tool for rapid and accurate identification of closely related species of M. guilliermondii complex and for differentiating emerging infectious yeasts of the Saccharomycotina CTG clade.
Figures


References
-
- Lachance MA, Boekhout T, Scorzetti G, Fell JW, Kurtzman CP. In: The Yeasts: A Taxonomic Study, Volume 2. 5. Kurtzman CP, Fell JW, Boekhout T, editor. San Diego: Elsevier; 2011. Candida Berkhout (1923) pp. 987–1278.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

