RBR E3 ubiquitin ligases: new structures, new insights, new questions
- PMID: 24576094
- PMCID: PMC3940038
- DOI: 10.1042/BJ20140006
RBR E3 ubiquitin ligases: new structures, new insights, new questions
Abstract
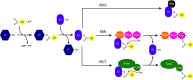
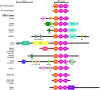
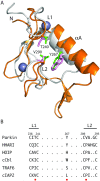
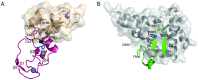
The RBR (RING-BetweenRING-RING) or TRIAD [two RING fingers and a DRIL (double RING finger linked)] E3 ubiquitin ligases comprise a group of 12 complex multidomain enzymes. This unique family of E3 ligases includes parkin, whose dysfunction is linked to the pathogenesis of early-onset Parkinson's disease, and HOIP (HOIL-1-interacting protein) and HOIL-1 (haem-oxidized IRP2 ubiquitin ligase 1), members of the LUBAC (linear ubiquitin chain assembly complex). The RBR E3 ligases share common features with both the larger RING and HECT (homologous with E6-associated protein C-terminus) E3 ligase families, directly catalysing ubiquitin transfer from an intrinsic catalytic cysteine housed in the C-terminal domain, as well as recruiting thioester-bound E2 enzymes via a RING domain. Recent three-dimensional structures and biochemical findings of the RBRs have revealed novel protein domain folds not previously envisioned and some surprising modes of regulation that have raised many questions. This has required renaming two of the domains in the RBR E3 ligases to more accurately reflect their structures and functions: the C-terminal Rcat (required-for-catalysis) domain, essential for catalytic activity, and a central BRcat (benign-catalytic) domain that adopts the same fold as the Rcat, but lacks a catalytic cysteine residue and ubiquitination activity. The present review discusses how three-dimensional structures of RBR (RING1-BRcat-Rcat) E3 ligases have provided new insights into our understanding of the biochemical mechanisms of these important enzymes in ubiquitin biology.
Figures

References
-
- Hershko A., Ciechanover A. The ubiquitin system. Annu. Rev. Biochem. 1998;67:425–479. - PubMed
-
- Deshaies R. J., Joazeiro C. A. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 2009;78:399–434. - PubMed
-
- Borden K. L., Freemont P. S. The RING finger domain: a recent example of a sequence-structure family. Curr. Opin. Struct. Biol. 1996;6:395–401. - PubMed
-
- Zheng N., Wang P., Jeffrey P. D., Pavletich N. P. Structure of a c-Cbl–UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell. 2000;102:533–539. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

