Specificity protein 4 (Sp4) regulates the transcription of AMPA receptor subunit GluA2 (Gria2)
- PMID: 24576410
- PMCID: PMC4024218
- DOI: 10.1016/j.bbamcr.2014.02.008
Specificity protein 4 (Sp4) regulates the transcription of AMPA receptor subunit GluA2 (Gria2)
Abstract
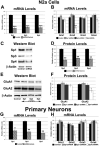
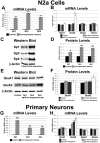
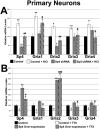
The alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptors are important glutamatergic receptors mediating fast excitatory synaptic transmission in the brain. The regulation of the four subunits of AMPA receptors, GluA1-4, is poorly understood. Excitatory synaptic transmission is highly energy-demanding, and this energy is derived mainly from the oxidative pathway. Recently, we found that specificity factor regulates all subunits of cytochrome c oxidase (COX), a critical energy-generating enzyme. COX is also regulated by nuclear respiratory factor 1 (NRF-1), which transcriptionally controls the Gria2 (GluA2) gene of AMPA receptors. The goal of the present study was to test our hypothesis that Sp-factors (Sp1, Sp3, and/or Sp4) also regulate AMPA subunit genes. If so, we wish to determine if Sp-factors and NRF-1 function via a complementary, concurrent and parallel, or a combination of complementary and concurrent/parallel mechanism. By means of multiple approaches, including electrophoretic mobility shift and supershift assays, chromatin immunoprecipitation, promoter mutations, real-time quantitative PCR, and western blot analysis, we found that Sp4, but not Sp1 or Sp3, regulates the Gria2, but not Gria1, 3, or 4, subunit gene of the AMPA receptor in a concurrent and parallel manner with NRF-1. Thus, Sp4 and NRF-1 both mediate the tight coupling between neuronal activity and energy metabolism at the transcriptional level.
Keywords: AMPA receptor; Gene regulation; GluA2; Sp4; Specificity protein 4; Transcription factor.
Copyright © 2014 Elsevier B.V. All rights reserved.
Conflict of interest statement
All authors have no conflict of interest to declare.
Figures





Similar articles
-
Specificity protein 4 functionally regulates the transcription of NMDA receptor subunits GluN1, GluN2A, and GluN2B.Biochim Biophys Acta. 2013 Dec;1833(12):2745-2756. doi: 10.1016/j.bbamcr.2013.07.002. Epub 2013 Jul 18. Biochim Biophys Acta. 2013. PMID: 23871830 Free PMC article.
-
Nuclear respiratory factor 2 regulates the transcription of AMPA receptor subunit GluA2 (Gria2).Biochim Biophys Acta. 2014 Dec;1843(12):3018-28. doi: 10.1016/j.bbamcr.2014.09.006. Epub 2014 Sep 22. Biochim Biophys Acta. 2014. PMID: 25245478 Free PMC article.
-
Specificity protein 4 (Sp4) transcriptionally regulates inhibitory GABAergic receptors in neurons.Biochim Biophys Acta. 2016 Jan;1863(1):1-9. doi: 10.1016/j.bbamcr.2015.10.005. Epub 2015 Oct 18. Biochim Biophys Acta. 2016. PMID: 26469128 Free PMC article.
-
Specificity Proteins (Sp) and Cancer.Int J Mol Sci. 2023 Mar 8;24(6):5164. doi: 10.3390/ijms24065164. Int J Mol Sci. 2023. PMID: 36982239 Free PMC article. Review.
-
AMPA receptors in schizophrenia: A systematic review of postmortem studies on receptor subunit expression and binding.Schizophr Res. 2022 May;243:98-109. doi: 10.1016/j.schres.2022.02.033. Epub 2022 Mar 3. Schizophr Res. 2022. PMID: 35247795
Cited by
-
Widespread promoter methylation of synaptic plasticity genes in long-term potentiation in the adult brain in vivo.BMC Genomics. 2017 Mar 23;18(1):250. doi: 10.1186/s12864-017-3621-x. BMC Genomics. 2017. PMID: 28335720 Free PMC article.
-
GlyT-1 Inhibition Attenuates Attentional But Not Learning or Motivational Deficits of the Sp4 Hypomorphic Mouse Model Relevant to Psychiatric Disorders.Neuropsychopharmacology. 2015 Nov;40(12):2715-26. doi: 10.1038/npp.2015.120. Epub 2015 Apr 24. Neuropsychopharmacology. 2015. PMID: 25907107 Free PMC article.
-
The Snail transcription factor CES-1 regulates glutamatergic behavior in C. elegans.PLoS One. 2021 Feb 2;16(2):e0245587. doi: 10.1371/journal.pone.0245587. eCollection 2021. PLoS One. 2021. PMID: 33529210 Free PMC article.
-
Genetic Diversity in Schizophrenia: Developmental Implications of Ultra-Rare, Protein-Truncating Mutations.Genes (Basel). 2024 Sep 17;15(9):1214. doi: 10.3390/genes15091214. Genes (Basel). 2024. PMID: 39336805 Free PMC article. Review.
-
Human stem cell-based models to study synaptic dysfunction and cognition in schizophrenia: A narrative review.Schizophr Res. 2024 Nov;273:78-97. doi: 10.1016/j.schres.2023.02.029. Epub 2023 Mar 14. Schizophr Res. 2024. PMID: 36925354 Review.
References
-
- Isaac JT, Ashby MC, McBain CJ. The role of the GluR2 subunit in AMPA receptor function and synaptic plasticity. Neuron. 2007;54:859–871. - PubMed
-
- Borges K, Dingledine R. AMPA receptors: molecular and functional diversity. Prog Brain Res. 1998;116:153–170. - PubMed
-
- Malinow R, Malenka RC. AMPA receptor trafficking and synaptic plasticity. Annu Rev Neurosci. 2002;25:103–126. - PubMed
-
- Shepherd JD, Huganir RL. The cell biology of synaptic plasticity: AMPA receptor trafficking. Annu Rev Cell Dev Biol. 2007;23:613–643. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

