A regulatory network of Drosophila germline stem cell self-renewal
- PMID: 24576427
- PMCID: PMC3998650
- DOI: 10.1016/j.devcel.2014.01.020
A regulatory network of Drosophila germline stem cell self-renewal
Abstract
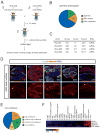
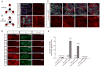
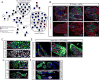
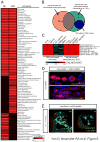
Stem cells possess the capacity to generate two cells of distinct fate upon division: one cell retaining stem cell identity and the other cell destined to differentiate. These cell fates are established by cell-type-specific genetic networks. To comprehensively identify components of these networks, we performed a large-scale RNAi screen in Drosophila female germline stem cells (GSCs) covering ∼25% of the genome. The screen identified 366 genes that affect GSC maintenance, differentiation, or other processes involved in oogenesis. Comparison of GSC regulators with neural stem cell self-renewal factors identifies common and cell-type-specific self-renewal genes. Importantly, we identify the histone methyltransferase Set1 as a GSC-specific self-renewal factor. Loss of Set1 in neural stem cells does not affect cell fate decisions, suggesting a differential requirement of H3K4me3 in different stem cell lineages. Altogether, our study provides a resource that will help to further dissect the networks underlying stem cell self-renewal.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

