Small regulatory RNAs from low-GC Gram-positive bacteria
- PMID: 24576839
- PMCID: PMC4152353
- DOI: 10.4161/rna.28036
Small regulatory RNAs from low-GC Gram-positive bacteria
Abstract
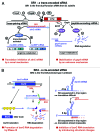
Small regulatory RNAs (sRNAs) that act by base-pairing were first discovered in so-called accessory DNA elements--plasmids, phages, and transposons--where they control replication, maintenance, and transposition. Since 2001, a huge body of work has been performed to predict and identify sRNAs in a multitude of bacterial genomes. The majority of chromosome-encoded sRNAs have been investigated in E. coli and other Gram-negative bacteria. However, during the past five years an increasing number of sRNAs were found in Gram-positive bacteria. Here, we outline our current knowledge on chromosome-encoded sRNAs from low-GC Gram-positive species that act by base-pairing, i.e., an antisense mechanism. We will focus on sRNAs with known targets and defined regulatory mechanisms with special emphasis on Bacillus subtilis.
Keywords: Bacillus subtilis; Streptococcus pneumoniae; base-pairing sRNA; low GC Gram-positive bacteria; small regulatory RNA.
Figures

References
-
- Fouquier d’Hérouel A, Wessner F, Halpern D, Ly-Vu J, Kennedy SP, Serror P, Aurell E, Repoila F. A simple and efficient method to search for selected primary transcripts: non-coding and antisense RNAs in the human pathogen Enterococcus faecalis. Nucleic Acids Res. 2011;39:e46. doi: 10.1093/nar/gkr012. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
