Detection of a recurrent DNAJB1-PRKACA chimeric transcript in fibrolamellar hepatocellular carcinoma
- PMID: 24578576
- PMCID: PMC4286414
- DOI: 10.1126/science.1249484
Detection of a recurrent DNAJB1-PRKACA chimeric transcript in fibrolamellar hepatocellular carcinoma
Abstract
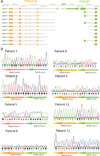
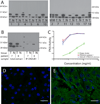
Fibrolamellar hepatocellular carcinoma (FL-HCC) is a rare liver tumor affecting adolescents and young adults with no history of primary liver disease or cirrhosis. We identified a chimeric transcript that is expressed in FL-HCC but not in adjacent normal liver and that arises as the result of a ~400-kilobase deletion on chromosome 19. The chimeric RNA is predicted to code for a protein containing the amino-terminal domain of DNAJB1, a homolog of the molecular chaperone DNAJ, fused in frame with PRKACA, the catalytic domain of protein kinase A. Immunoprecipitation and Western blot analyses confirmed that the chimeric protein is expressed in tumor tissue, and a cell culture assay indicated that it retains kinase activity. Evidence supporting the presence of the DNAJB1-PRKACA chimeric transcript in 100% of the FL-HCCs examined (15/15) suggests that this genetic alteration contributes to tumor pathogenesis.
Figures

References
-
- El-Serag HB, Davila JA. Is fibrolamellar carcinoma different from hepatocellular carcinoma? A US population-based study. 2004;39:798–803. - PubMed
-
- Edmondson HA. Differential diagnosis of tumors and tumor-like lesions of liver in infancy and childhood. 1956;91:168–186. - PubMed
-
- Torbenson M. Review of the clinicopathologic features of fibrolamellar carcinoma. 2007;14:217–223. - PubMed
-
- Craig JR, Peters RL, Edmondson HA, Omata M. Fibrolamellar carcinoma of the liver: a tumor of adolescents and young adults with distinctive clinico-pathologic features. 1980;46:372–379. - PubMed
-
- Stipa F, et al. Outcome of patients with fibrolamellar hepatocellular carcinoma. 2006;106:1331–1338. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

