New insights into bacterial chemoreceptor array structure and assembly from electron cryotomography
- PMID: 24580139
- PMCID: PMC3985956
- DOI: 10.1021/bi5000614
New insights into bacterial chemoreceptor array structure and assembly from electron cryotomography
Erratum in
- Biochemistry. 2014 Oct 21;53(41):6624
Abstract
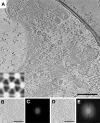
Bacterial chemoreceptors cluster in highly ordered, cooperative, extended arrays with a conserved architecture, but the principles that govern array assembly remain unclear. Here we show images of cellular arrays as well as selected chemoreceptor complexes reconstituted in vitro that reveal new principles of array structure and assembly. First, in every case, receptors clustered in a trimers-of-dimers configuration, suggesting this is a highly favored fundamental building block. Second, these trimers-of-receptor dimers exhibited great versatility in the kinds of contacts they formed with each other and with other components of the signaling pathway, although only one architectural type occurred in native arrays. Third, the membrane, while it likely accelerates the formation of arrays, was neither necessary nor sufficient for lattice formation. Molecular crowding substituted for the stabilizing effect of the membrane and allowed cytoplasmic receptor fragments to form sandwiched lattices that strongly resemble the cytoplasmic chemoreceptor arrays found in some bacterial species. Finally, the effective determinant of array structure seemed to be CheA and CheW, which formed a "superlattice" of alternating CheA-filled and CheA-empty rings that linked receptor trimers-of-dimer units into their native hexagonal lattice. While concomitant overexpression of receptors, CheA, and CheW yielded arrays with native spacing, the CheA occupancy was lower and less ordered, suggesting that temporal and spatial coordination of gene expression driven by a single transcription factor may be vital for full order, or that array overgrowth may trigger a disassembly process. The results described here provide new insights into the assembly intermediates and assembly mechanism of this massive macromolecular complex.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

