Modular assembly of designer PUF proteins for specific post-transcriptional regulation of endogenous RNA
- PMID: 24581042
- PMCID: PMC3943411
- DOI: 10.1186/1754-1611-8-7
Modular assembly of designer PUF proteins for specific post-transcriptional regulation of endogenous RNA
Abstract
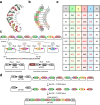
Background: Due to their modular repeat structure, Pumilio/fem-3 mRNA binding factor (PUF) proteins are promising candidates for designer RNA-binding protein (RBP) engineering. To further facilitate the application of the PUF domain for the sequence-specific RBP engineering, a rapid cloning approach is desirable that would allow efficient introduction of multiple key amino acid mutations in the protein. Here, we report the implementation of the Golden Gate cloning method for an efficient one-step assembly of a designer PUF domain for RNA specificity engineering.
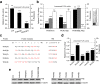
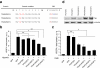
Results: We created a repeat module library that is potentially capable of generating a PUF domain with any desired specificity. PUF domains with multiple repeat modifications for the recognition of altered RNA targets were obtained in a one-step assembly reaction, which was found to be highly efficient. The new PUF variants exhibited high in vitro binding efficiencies to cognate RNA sequences, corroborating the applicability of the modular approach for PUF engineering. To demonstrate the application of the PUF domain assembly method for RBP engineering, we fused the PUF domain to a post-transcriptional regulator and observed a sequence-specific reporter and endogenous gene repression in human cell lines.
Conclusions: The Golden Gate based cloning approach thus should allow greater flexibility and speed in implementing the PUF protein scaffold for engineering designer RBPs, and facilitate its use as a tool in basic and applied biology and medicine.
Figures
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

