Nuclear pores protect genome integrity by assembling a premitotic and Mad1-dependent anaphase inhibitor
- PMID: 24581499
- PMCID: PMC3947552
- DOI: 10.1016/j.cell.2014.01.010
Nuclear pores protect genome integrity by assembling a premitotic and Mad1-dependent anaphase inhibitor
Abstract
The spindle assembly checkpoint (SAC) delays anaphase until all chromosomes are bioriented on the mitotic spindle. Under current models, unattached kinetochores transduce the SAC by catalyzing the intramitotic production of a diffusible inhibitor of APC/C(Cdc20) (the anaphase-promoting complex/cyclosome and its coactivator Cdc20, a large ubiquitin ligase). Here we show that nuclear pore complexes (NPCs) in interphase cells also function as scaffolds for anaphase-inhibitory signaling. This role is mediated by Mad1-Mad2 complexes tethered to the nuclear basket, which activate soluble Mad2 as a binding partner and inhibitor of Cdc20 in the cytoplasm. Displacing Mad1-Mad2 from nuclear pores accelerated anaphase onset, prevented effective correction of merotelic errors, and increased the threshold of kinetochore-dependent signaling needed to halt mitosis in response to spindle poisons. A heterologous Mad1-NPC tether restored Cdc20 inhibitor production and normal M phase control. We conclude that nuclear pores and kinetochores both emit "wait anaphase" signals that preserve genome integrity.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures
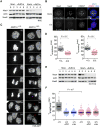
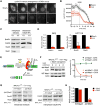
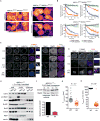



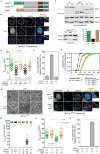
Comment in
-
Nuclear pores set the speed limit for mitosis.Cell. 2014 Feb 27;156(5):868-9. doi: 10.1016/j.cell.2014.02.004. Cell. 2014. PMID: 24581486 Free PMC article.
-
Mitosis: a safety net for successful mitosis.Nat Rev Mol Cell Biol. 2014 Apr;15(4):220-1. doi: 10.1038/nrm3777. Nat Rev Mol Cell Biol. 2014. PMID: 24651536 No abstract available.
References
-
- Arnaoutov A, Azuma Y, Ribbeck K, Joseph J, Boyarchuk Y, Karpova T, McNally J, Dasso M. Crm1 is a mitotic effector of Ran-GTP in somatic cells. Nat Cell Biol. 2005;7:626–632. - PubMed
-
- Berdougo E, Terret M, Jallepalli P. Functional dissection of mitotic regulators through gene targeting in human somatic cells. Methods Mol Biol. 2009;545:21–37. - PubMed
-
- Cairo LV, Ptak C, Wozniak RW. Mitosis-Specific Regulation of Nuclear Transport by the Spindle Assembly Checkpoint Protein Mad1p. Mol Cell 2012 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

