Structural organisation of the type IV secretion systems
- PMID: 24581689
- PMCID: PMC3969286
- DOI: 10.1016/j.mib.2013.11.001
Structural organisation of the type IV secretion systems
Abstract
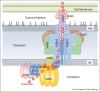
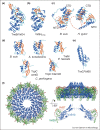
Type IV secretion (T4S) systems are large dynamic nanomachines that transport DNAs and/or proteins through the membranes of bacteria. Because of their complexity and multi-protein organisation, T4S systems have been extremely challenging to study structurally. However in the past five years significant milestones have been achieved by X-ray crystallography and cryo-electron microscopy. This review describes some of the more recent advances: the structures of some of the protein components of the T4S systems and the complete core complex structure that was determined using electron microscopy.
Copyright © 2013 The Authors. Published by Elsevier Ltd.. All rights reserved.
Figures


Similar articles
-
Structure of a bacterial type IV secretion core complex at subnanometre resolution.EMBO J. 2013 Apr 17;32(8):1195-204. doi: 10.1038/emboj.2013.58. Epub 2013 Mar 19. EMBO J. 2013. PMID: 23511972 Free PMC article.
-
Type IV secretion machinery: molecular architecture and function.Biochem Soc Trans. 2013 Feb 1;41(1):17-28. doi: 10.1042/BST20120332. Biochem Soc Trans. 2013. PMID: 23356253 Review.
-
Bacterial type IV secretion systems in human disease.Mol Microbiol. 2009 Jul;73(2):141-51. doi: 10.1111/j.1365-2958.2009.06751.x. Epub 2009 Jun 8. Mol Microbiol. 2009. PMID: 19508287 Free PMC article. Review.
-
Molecular architecture of bacterial type IV secretion systems.Trends Biochem Sci. 2010 Dec;35(12):691-8. doi: 10.1016/j.tibs.2010.06.002. Epub 2010 Jul 10. Trends Biochem Sci. 2010. PMID: 20621482 Review.
-
Cryo-EM structure of the bacteria-killing type IV secretion system core complex from Xanthomonas citri.Nat Microbiol. 2018 Dec;3(12):1429-1440. doi: 10.1038/s41564-018-0262-z. Epub 2018 Oct 22. Nat Microbiol. 2018. PMID: 30349081 Free PMC article.
Cited by
-
Architecture of the outer-membrane core complex from a conjugative type IV secretion system.Nat Commun. 2021 Nov 25;12(1):6834. doi: 10.1038/s41467-021-27178-8. Nat Commun. 2021. PMID: 34824240 Free PMC article.
-
Arabidopsis RETICULON-LIKE3 (RTNLB3) and RTNLB8 Participate in Agrobacterium-Mediated Plant Transformation.Int J Mol Sci. 2018 Feb 24;19(2):638. doi: 10.3390/ijms19020638. Int J Mol Sci. 2018. PMID: 29495267 Free PMC article.
-
Xanthomonas spp. Infecting Araceae and Araliaceae: Taxonomy, Phylogeny, and Potential Virulence Mechanisms.Biology (Basel). 2025 Jun 25;14(7):766. doi: 10.3390/biology14070766. Biology (Basel). 2025. PMID: 40723327 Free PMC article. Review.
-
Comparative Genome Analysis of Two Isolates of the Fish Pathogen Piscirickettsia salmonis from Different Hosts Reveals Major Differences in Virulence-Associated Secretion Systems.Genome Announc. 2014 Dec 18;2(6):e01219-14. doi: 10.1128/genomeA.01219-14. Genome Announc. 2014. PMID: 25523762 Free PMC article.
-
Draft genome sequence for virulent and avirulent strains of Xanthomonas arboricola isolated from Prunus spp. in Spain.Stand Genomic Sci. 2016 Jan 28;11:12. doi: 10.1186/s40793-016-0132-3. eCollection 2016. Stand Genomic Sci. 2016. PMID: 26823958 Free PMC article.
References
-
- Omori K., Idei A. Gram-negative bacterial ATP-binding cassette protein exporter family and diverse secretory proteins. J Biosci Bioeng. 2003;95:1–12. - PubMed
-
- Filloux A. The underlying mechanisms of type II protein secretion. Biochim Biophys Acta. 2004;1694:163–179. - PubMed
-
- Cornelis G.R., Van Gijsegem F. Assembly and function of type III secretory systems. Annu Rev Microbiol. 2000;54:735–774. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

