Discovery of unconventional kinetochores in kinetoplastids
- PMID: 24582333
- PMCID: PMC3978658
- DOI: 10.1016/j.cell.2014.01.049
Discovery of unconventional kinetochores in kinetoplastids
Abstract
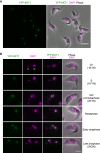
The kinetochore is the macromolecular protein complex that directs chromosome segregation in eukaryotes. It has been widely assumed that the core kinetochore consists of proteins that are common to all eukaryotes. However, no conventional kinetochore components have been identified in any kinetoplastid genome, thus challenging this assumption of universality. Here, we report the identification of 19 kinetochore proteins (KKT1-19) in Trypanosoma brucei. The majority is conserved among kinetoplastids, but none of them has detectable homology to conventional kinetochore proteins. These proteins instead have a variety of features not found in conventional kinetochore proteins. We propose that kinetoplastids build kinetochores using a distinct set of proteins. These findings provide important insights into the longstanding problem of the position of the root of the eukaryotic tree of life.
Copyright © 2014 The Authors. Published by Elsevier Inc. All rights reserved.
Figures









Comment in
-
Cell division: the prehistorichore?Curr Biol. 2014 Jun 2;24(11):R529-32. doi: 10.1016/j.cub.2014.04.035. Curr Biol. 2014. PMID: 24892916
References
-
- Allen J.W.A., Ferguson S.J., Ginger M.L. Distinctive biochemistry in the trypanosome mitochondrial intermembrane space suggests a model for stepwise evolution of the MIA pathway for import of cysteine-rich proteins. FEBS Lett. 2008;582:2817–2825. - PubMed
Supplemental References
-
- Cheeseman I.M., Chappie J.S., Wilson-Kubalek E.M., Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. - PubMed
-
- Redmond S., Vadivelu J., Field M.C. RNAit: an automated web-based tool for the selection of RNAi targets in Trypanosoma brucei. Mol. Biochem. Parasitol. 2003;128:115–118. - PubMed
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

