(19)F NMR reveals multiple conformations at the dimer interface of the nonstructural protein 1 effector domain from influenza A virus
- PMID: 24582435
- PMCID: PMC4110948
- DOI: 10.1016/j.str.2014.01.010
(19)F NMR reveals multiple conformations at the dimer interface of the nonstructural protein 1 effector domain from influenza A virus
Abstract
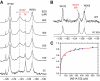
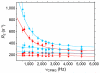
Nonstructural protein 1 of influenza A virus (NS1A) is a conserved virulence factor comprised of an N-terminal double-stranded RNA (dsRNA)-binding domain and a multifunctional C-terminal effector domain (ED), each of which can independently form symmetric homodimers. Here we apply (19)F NMR to NS1A from influenza A/Udorn/307/1972 virus (H3N2) labeled with 5-fluorotryptophan, and we demonstrate that the (19)F signal of Trp187 is a sensitive, direct monitor of the ED helix:helix dimer interface. (19)F relaxation dispersion data reveal the presence of conformational dynamics within this functionally important protein:protein interface, whose rate is more than three orders of magnitude faster than the kinetics of ED dimerization. (19)F NMR also affords direct spectroscopic evidence that Trp187, which mediates intermolecular ED:ED interactions required for cooperative dsRNA binding, is solvent exposed in full-length NS1A at concentrations below aggregation. These results have important implications for the diverse roles of this NS1A epitope during influenza virus infection.
Copyright © 2014 Elsevier Ltd. All rights reserved.
Figures




References
-
- Ahmed AH, Loh AP, Jane DE, Oswald RE. Dynamics of the S1S2 glutamate binding domain of GluR2 measured using 19F NMR spectroscopy. J Biol Chem. 2007;282:12773–12784. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

