Bat-derived influenza-like viruses H17N10 and H18N11
- PMID: 24582528
- PMCID: PMC7127364
- DOI: 10.1016/j.tim.2014.01.010
Bat-derived influenza-like viruses H17N10 and H18N11
Abstract
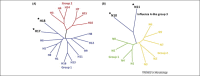
Shorebirds and waterfowls are believed to be the reservoir hosts for influenza viruses, whereas swine putatively act as mixing vessels. The recent identification of two influenza-like virus genomes (designated H17N10 and H18N11) from bats has challenged this notion. A crucial question concerns the role bats might play in influenza virus ecology. Structural and functional studies of the two major surface envelope proteins, hemagglutinin (HA) and neuraminidase (NA), demonstrate that neither has canonical HA or NA functions found in influenza viruses. However, putative functional modules and domains in other encoded proteins are conserved, and the N-terminal domain of the H17N10 polymerase subunit PA has a classical structure and function. Therefore, potential genomic reassortments of such influenza-like viruses with canonical influenza viruses cannot be excluded at this point and should be assessed.
Keywords: H17N10; H18N11; PA; bat-derived influenza-like virus; hemagglutinin (HA); neuraminidase (NA); reassortment.
Copyright © 2014 Elsevier Ltd. All rights reserved.
Figures



References
-
- Palese P., Shaw M.L. Orthomyxoviridae: the viruses and their replication. In: Knipe D.M., Howley P.M., editors. Fields Virology. 4th edn. Lippincott Williams & Wilkins; 2007. pp. 1647–1690.
-
- Cox N.J., Subbarao K. Global epidemiology of influenza: past and present. Annu. Rev. Med. 2000;51:407–421. - PubMed
-
- Kobasa D. Enhanced virulence of influenza A viruses with the haemagglutinin of the 1918 pandemic virus. Nature. 2004;431:703–707. - PubMed
-
- Novel Swine-Origin Influenza A (H1N1) Virus Investigation Team Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N. Engl. J. Med. 2009;360:2605–2615. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

