Novel primate miRNAs coevolved with ancient target genes in germinal zone-specific expression patterns
- PMID: 24583023
- PMCID: PMC4020629
- DOI: 10.1016/j.neuron.2014.01.017
Novel primate miRNAs coevolved with ancient target genes in germinal zone-specific expression patterns
Abstract
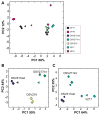
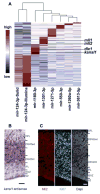
Major nonprimate-primate differences in cortico-genesis include the dimensions, precursor lineages, and developmental timing of the germinal zones (GZs). microRNAs (miRNAs) of laser-dissected GZ compartments and cortical plate (CP) from embryonic E80 macaque visual cortex were deep sequenced. The CP and the GZ including ventricular zone (VZ) and outer and inner subcompartments of the outer subventricular zone (OSVZ) in area 17 displayed unique miRNA profiles. miRNAs present in primate, but absent in rodent, contributed disproportionately to the differential expression between GZ subregions. Prominent among the validated targets of these miRNAs were cell-cycle and neurogenesis regulators. Coevolution between the emergent miRNAs and their targets suggested that novel miRNAs became integrated into ancient gene circuitry to exert additional control over proliferation. We conclude that multiple cell-cycle regulatory events contribute to the emergence of primate-specific cortical features, including the OSVZ, generated enlarged supragranular layers, largely responsible for the increased primate cortex computational abilities.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures

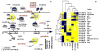
References
-
- Anderson SA, Eisenstat DD, Shi L, Rubenstein LR. Interneuron migration from basal forebrain to neocortex: Dependence on Dlx genes. Science. 1997;278:474–476. - PubMed
-
- Betizeau M, Cortay V, Patti D, Pfister S, Gautier E, Bellemin-Menard A, Afanassieff M, Huissoud C, Douglas RJ, Kennedy H, Dehay C. Precursor diversity and complexity of lineage relationships in the outer subventricular zone (OSVZ) of the primate. Neuron. 2013;80:442–457. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

