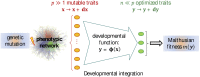
Fisher's geometrical model emerges as a property of complex integrated phenotypic networks
- PMID: 24583582
- PMCID: PMC4012483
- DOI: 10.1534/genetics.113.160325
Fisher's geometrical model emerges as a property of complex integrated phenotypic networks
Abstract
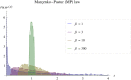
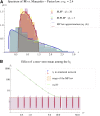
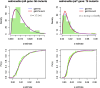
Models relating phenotype space to fitness (phenotype-fitness landscapes) have seen important developments recently. They can roughly be divided into mechanistic models (e.g., metabolic networks) and more heuristic models like Fisher's geometrical model. Each has its own drawbacks, but both yield testable predictions on how the context (genomic background or environment) affects the distribution of mutation effects on fitness and thus adaptation. Both have received some empirical validation. This article aims at bridging the gap between these approaches. A derivation of the Fisher model "from first principles" is proposed, where the basic assumptions emerge from a more general model, inspired by mechanistic networks. I start from a general phenotypic network relating unspecified phenotypic traits and fitness. A limited set of qualitative assumptions is then imposed, mostly corresponding to known features of phenotypic networks: a large set of traits is pleiotropically affected by mutations and determines a much smaller set of traits under optimizing selection. Otherwise, the model remains fairly general regarding the phenotypic processes involved or the distribution of mutation effects affecting the network. A statistical treatment and a local approximation close to a fitness optimum yield a landscape that is effectively the isotropic Fisher model or its extension with a single dominant phenotypic direction. The fit of the resulting alternative distributions is illustrated in an empirical data set. These results bear implications on the validity of Fisher's model's assumptions and on which features of mutation fitness effects may vary (or not) across genomic or environmental contexts.
Keywords: Fisher’s geometrical model; mutation fitness effects; network biology; random matrix theory; systems biology.
Figures


Similar articles
-
The fitness effect of mutations across environments: Fisher's geometrical model with multiple optima.Evolution. 2015 Jun;69(6):1433-1447. doi: 10.1111/evo.12671. Epub 2015 Jun 10. Evolution. 2015. PMID: 25908434
-
Fisher's geometrical model of fitness landscape and variance in fitness within a changing environment.Evolution. 2012 Aug;66(8):2350-68. doi: 10.1111/j.1558-5646.2012.01610.x. Epub 2012 Apr 13. Evolution. 2012. PMID: 22834737
-
Genotypic Complexity of Fisher's Geometric Model.Genetics. 2017 Jun;206(2):1049-1079. doi: 10.1534/genetics.116.199497. Epub 2017 Apr 26. Genetics. 2017. PMID: 28450460 Free PMC article.
-
A general multivariate extension of Fisher's geometrical model and the distribution of mutation fitness effects across species.Evolution. 2006 May;60(5):893-907. Evolution. 2006. PMID: 16817531 Review.
-
Effects of new mutations on fitness: insights from models and data.Ann N Y Acad Sci. 2014 Jul;1320(1):76-92. doi: 10.1111/nyas.12460. Epub 2014 May 30. Ann N Y Acad Sci. 2014. PMID: 24891070 Free PMC article. Review.
Cited by
-
Diminishing-returns epistasis among random beneficial mutations in a multicellular fungus.Proc Biol Sci. 2016 Aug 31;283(1837):20161376. doi: 10.1098/rspb.2016.1376. Proc Biol Sci. 2016. PMID: 27559062 Free PMC article.
-
Coadapted genomes and selection on hybrids: Fisher's geometric model explains a variety of empirical patterns.Evol Lett. 2018 Aug 14;2(5):472-498. doi: 10.1002/evl3.66. eCollection 2018 Oct. Evol Lett. 2018. PMID: 30283696 Free PMC article.
-
Genetic Paths to Evolutionary Rescue and the Distribution of Fitness Effects Along Them.Genetics. 2020 Feb;214(2):493-510. doi: 10.1534/genetics.119.302890. Epub 2019 Dec 10. Genetics. 2020. PMID: 31822480 Free PMC article.
-
Inference of the Distribution of Selection Coefficients for New Nonsynonymous Mutations Using Large Samples.Genetics. 2017 May;206(1):345-361. doi: 10.1534/genetics.116.197145. Epub 2017 Mar 1. Genetics. 2017. PMID: 28249985 Free PMC article.
-
The Utility of Fisher's Geometric Model in Evolutionary Genetics.Annu Rev Ecol Evol Syst. 2014 Nov 1;45:179-201. doi: 10.1146/annurev-ecolsys-120213-091846. Annu Rev Ecol Evol Syst. 2014. PMID: 26740803 Free PMC article.
References
-
- Bai Z., Silverstein J. W., 2010. Spectral Analysis of Large Dimensional Random Matrices, Ed. 2. now Publishers Inc., Hanover, MA.
-
- Barabasi A. L., Oltvai Z. N., 2004. Network biology: understanding the cell’s functional organization. Nat. Rev. Genet. 5: 101–113. - PubMed
-
- Barton N. H., Coe J. B., 2009. On the application of statistical physics to evolutionary biology. J. Theor. Biol. 259: 317–324. - PubMed
-
- Bataillon T., 2000. Estimation of spontaneous genome-wide mutation rate parameters: Whither beneficial mutations? Heredity 84: 497–501. - PubMed
-
- Baxter G. J., Blythe R. A., McKane A. J., 2007. Exact solution of the multi-allelic diffusion model. Math. Biosci. 209: 124–170. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

