Notch1 regulates progenitor cell proliferation and differentiation during mouse yolk sac hematopoiesis
- PMID: 24583642
- PMCID: PMC4207476
- DOI: 10.1038/cdd.2014.27
Notch1 regulates progenitor cell proliferation and differentiation during mouse yolk sac hematopoiesis
Abstract
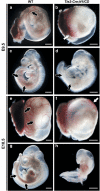
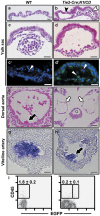
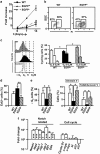
Loss-of-function studies have demonstrated the essential role of Notch in definitive embryonic mouse hematopoiesis. We report here the consequences of Notch gain-of-function in mouse embryo hematopoiesis, achieved by constitutive expression of Notch1 intracellular domain (N1ICD) in angiopoietin receptor tyrosine kinase receptor-2 (Tie2)-derived enhanced green fluorescence protein (EGFP(+)) hematovascular progenitors. At E9.5, N1ICD expression led to the absence of the dorsal aorta hematopoietic clusters and of definitive hematopoiesis. The EGFP(+) transient multipotent progenitors, purified from E9.5 to 10.5 Tie2-Cre;N1ICD yolk sac (YS) cells, had strongly reduced hematopoietic potential, whereas they had increased numbers of hemogenic endothelial cells. Late erythroid cell differentiation stages and mature myeloid cells (Gr1(+), MPO(+)) were also strongly decreased. In contrast, EGFP(+) erythro-myeloid progenitors, immature and intermediate differentiation stages of YS erythroid and myeloid cell lineages, were expanded. Tie2-Cre;N1ICD YS had reduced numbers of CD41(++) megakaryocytes, and these produced reduced below-normal numbers of immature colonies in vitro and their terminal differentiation was blocked. Cells from Tie2-Cre;N1ICD YS had a higher proliferation rate and lower apoptosis than wild-type (WT) YS cells. Quantitative gene expression analysis of FACS-purified EGFP(+) YS progenitors revealed upregulation of Notch1-related genes and alterations in genes involved in hematopoietic differentiation. These results represent the first in vivo evidence of a role for Notch signaling in YS transient definitive hematopoiesis. Our results show that constitutive Notch1 activation in Tie2(+) cells hampers YS hematopoiesis of E9.5 embryos and demonstrate that Notch signaling regulates this process by balancing the proliferation and differentiation dynamics of lineage-restricted intermediate progenitors.
Figures




Similar articles
-
Cardiovascular and hematopoietic defects associated with Notch1 activation in embryonic Tie2-expressing populations.Circ Res. 2008 Aug 15;103(4):423-31. doi: 10.1161/CIRCRESAHA.108.177808. Epub 2008 Jul 10. Circ Res. 2008. PMID: 18617694 Free PMC article.
-
Multiple cell populations generate macrophage progenitors in the early yolk sac.Cell Mol Life Sci. 2022 Feb 27;79(3):159. doi: 10.1007/s00018-022-04203-7. Cell Mol Life Sci. 2022. PMID: 35224692 Free PMC article.
-
LYVE1 Marks the Divergence of Yolk Sac Definitive Hemogenic Endothelium from the Primitive Erythroid Lineage.Cell Rep. 2016 Nov 22;17(9):2286-2298. doi: 10.1016/j.celrep.2016.10.080. Cell Rep. 2016. PMID: 27880904 Free PMC article.
-
Embryonic hematopoiesis.Blood Cells Mol Dis. 2013 Dec;51(4):226-31. doi: 10.1016/j.bcmd.2013.08.004. Epub 2013 Sep 13. Blood Cells Mol Dis. 2013. PMID: 24041595 Review.
-
Hematopoietic stem cell-independent hematopoiesis: emergence of erythroid, megakaryocyte, and myeloid potential in the mammalian embryo.FEBS Lett. 2016 Nov;590(22):3965-3974. doi: 10.1002/1873-3468.12459. Epub 2016 Oct 27. FEBS Lett. 2016. PMID: 27790707 Review.
Cited by
-
A NOTCH4 intron 28 regulatory element controls arterial specification and lymphoid development from hPSCs.Blood Vessel Thromb Hemost. 2025 Jan 15;2(2):100046. doi: 10.1016/j.bvth.2025.100046. eCollection 2025 May. Blood Vessel Thromb Hemost. 2025. PMID: 40766274 Free PMC article.
-
Loss of Apela Peptide in Mice Causes Low Penetrance Embryonic Lethality and Defects in Early Mesodermal Derivatives.Cell Rep. 2017 Aug 29;20(9):2116-2130. doi: 10.1016/j.celrep.2017.08.014. Cell Rep. 2017. PMID: 28854362 Free PMC article.
-
Evaluation of velvet antler total protein effect on bone marrow‑derived endothelial progenitor cells.Mol Med Rep. 2017 Sep;16(3):3161-3168. doi: 10.3892/mmr.2017.7019. Epub 2017 Jul 15. Mol Med Rep. 2017. PMID: 28714033 Free PMC article.
-
Three-Dimensional Environment Sustains Hematopoietic Stem Cell Differentiation into Platelet-Producing Megakaryocytes.PLoS One. 2015 Aug 27;10(8):e0136652. doi: 10.1371/journal.pone.0136652. eCollection 2015. PLoS One. 2015. PMID: 26313154 Free PMC article.
-
Identification and characterization of enhancer elements controlling cell type-specific and signalling dependent chromatin programming during hematopoietic development.Stem Cell Investig. 2023 Jun 25;10:14. doi: 10.21037/sci-2023-011. eCollection 2023. Stem Cell Investig. 2023. PMID: 37404470 Free PMC article. Review.
References
-
- Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. - PubMed
-
- Radtke F, Fasnacht N, Macdonald HR. Notch signalling in the immune system. Immunity. 2010;32:14–27. - PubMed
-
- Lieber T, Kidd S, Alcamo E, Corbin V, Young MW. Antineurogenic phenotypes induced by truncated Notch proteins indicate a role in signal transduction and may point to a novel function for Notch in nuclei. Genes Dev. 1993;7:1949–1965. - PubMed
-
- Rebay I, Fehon RG, Artavanis-Tsakonas S. Specific truncations of Drosophila Notch define dominant activated and dominant negative forms of the receptor. Cell. 1993;74:319–329. - PubMed
-
- Struhl G, Fitzgerald K, Greenwald I. Intrinsic activity of the Lin-12 and Notch intracellular domains in vivo. Cell. 1993;74:331–345. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

